Oral Presentation Abstracts
Amalie Skogvold
Amalie Skogvold1, Heidi Therese Hillier1, Ingar Leiros1
1 Department of Chemistry, UiT – The Arctic University of Norway
ω-transaminases are biocatalysts highly sought after in the pharmaceutical industry, as a green alternative for production of valuable chiral amines. The DABA ω-transaminases EctB and DoeD catalyze the forward and reverse transaminase reaction in the bacterial Ectoine biosynthesis and degradation pathway, respectively. Ectoine is a highly valuable compound used both in the cosmetics and pharmaceutical industry due to its many novel properties such as cell protectant, protein stabilizer, skin and UV protectant. It is produced mainly by a method called bacterial milking, but more recent research is done on heterologous production using non-halophilic bacteria such as Escherichia coli. Despite the high marked demand of Ectoine, there is little research done on characterizing the enzymes in the Ectoine biosynthesis or degradation pathway. Recently, we solved the first crystal structure of DoeD, and completed a biochemical and biophysical characterization of the transaminase, including exploring the substrate scope for other potential uses of the enzyme as a pharmaceutical biocatalyst. Previous work by the Ectoine Research group includes solving the crystal structure of EctB from the model organism for Ectoine production Chromohalobacter salexigens (Hillier et al., 2020). The group aims to characterize all the core enzymes that synthesize Ectoine, including EctB, EctA and EctC, as well as DoeD which is a part of the Ectoine degradation pathway. Further work will also include rational design to improve operational stability and efficiency of especially the transaminases EctB and DoeD.
References:
- Hillier, H. T., Altermark, B., & Leiros, I. (2020). The crystal structure of the tetrameric DABA‐aminotransferase EctB, a rate‐limiting enzyme in the ectoine biosynthesis pathway. The FEBS journal, 287(21), 4641-4658.
Antonia Areali
Antonia Areali1
1 Norwegian University of Life Sciences
Secretion systems in bacteria are of high scientific importance, as various molecules can be secreted from bacteria to respond in different processes. Many factors can affect bacterial secretion systems and the export of proteins, therefore understanding of those factors is the key for a proper biotechnological exploitation of microbial secretion systems. Currently, several bacteria workhorses like Escherichia coli have been extensively studied and engineered to produce high value proteins. The purpose of this research is to study, engineer, optimize and exploit the flagellar secretion systems T3SS which is one of the most important secretion systems in bacteria and has shown a remarkably fast exportation of proteins to the medium. As proof-of-concept, within this project we will establish biotechnological production of high value proteins as, for instance, protein-based fish vaccines at different production scales.
Ezgi Başavcı
Ezgi Basavci1, Alvja Mali3, Erik Agner1, Mangala Srinivas3, Magne Olav Sydnes2
1 Polypure AS, Norway,
2 University of Stavanger, Norway
3 Wageningen University and Research, The Netherlands
Magnetic resonance imaging (MRI) is a high-resolution and non-invasive preclinical and clinical imaging method using a strong magnetic field.1 In this project we aim to develop a highly reproducible novel contrast agent (CA) for 19F MRI and study its physiochemical and biological properties. Since the 19F nucleus is extremely sensitive for MRI and the 19F signal is directly proportional to the amount of 19F present in the fluorine-based CAs, such as perfluoro-15-crown-5-ether (PFCE), making it possible to quantify precisely from the image, unlike conventional gadolinium (Gd3+) based CAs that alter the existing proton signal. PFCEs possess a substantial amount of 19F nuclei, but they are immiscible with lipophilic and hydrophilic solvents.
In this study, poly (lactic-co-glycolic acid) (PLGA), a biodegradable and biocompatible polyester approved by and Drug Administration (FDA), is used as a versatile nanocarrier to encapsulate PFCE. However, hydrophobic PLGA polymer and PFCEs have some drawbacks as low clearance by the reticuloendothelial system (RES), and long half-life. Srinivas and co-workers reported that PFCE domains are stabilized by the polymeric matrix as a multicore structure which is extremely advantageous in terms of 15-fold faster removal from the body and resulting in a half-life of 16 days.2 It has been proposed that attaching monodisperse and structurally well-defined polyethylene glycol (PEG) derivatives onto the surface of nanoparticles enhance biocompatibility and circumvent the opsonins.3 We are focusing on the reproducibility of nanoparticle production securing the same pharmaceutical quality between batches.
Currently, we are synthesizing and purifying heterobifunctional PEG amines followed by attaching them to the PFCE encapsulated PLGA nanoparticle. Physiochemical characterization of each product has been done by FTIR, DLS, Zeta, 1H NMR, and 19F MRI. The effect of PEG length and end group functionality on nanoparticles for modulating biodistribution and uptake by target cells will be investigated. It is likely that further experiments will demonstrate the importance of oligomer purity and the impact of the heterobifunctional PEG design.
References
- Ruiz-Cabello, J., Barnett, B. P., Bottomley, P. A. & Bulte, J. W. M. Fluorine (19F) MRS and MRI in biomedicine. NMR in Biomedicine vol. 24 114–129 (2011).
- Koshkina, O. et al. Multicore Liquid Perfluorocarbon-Loaded Multimodal Nanoparticles for Stable Ultrasound and 19F MRI Applied to In Vivo Cell Tracking. Advanced Functional Materials 29, 110770, (2019).
- Gajbhiye, K. R., Pawar, A., Mahadik, K. R. & Gajbhiye, V. PEGylated nanocarriers: A promising tool for targeted delivery to the brain. Colloids and Surfaces B: Biointerfaces vol. 187,114-129, (2020).
This work has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Actions grant agreement No 859908
Jan Benedict Spannenkrebs
Jan Benedict Spannenkrebs1, Marianna Karava2, Johannes Kabisch1
1 Department of Biotechnology and Food Science, Norwegian University of Science and Technology, Trondheim
2 Institute of Molecular Biotechnology, Graz University of Technology, Austria
Although enzymes nowadays are often used to reduce the demand a certain process has in terms of energy, resources and water, the current large-scale production of enzymes, especially purified ones remains a significant consumer of said limited commodities1.
The Gram-positive bacterium Bacillus subtilis is widely used in biotechnology due to the ease of genome manipulation, due to being a good secreter of enzymes and many processes with B. subtilis having obtained a generally recognized as safe (GRAS) status from the FDA. In case of worsening environmental conditions, Bacillus subtilis cells can sporulate, safeguarding their DNA in a spore highly resistant to environmental stress.
The possibility of displaying enzymes on these bacterial endospores, is a method to combine enzyme production and immobilization into a single process2. This display usually involves the creation of a genetically linked fusion protein, which consists of a protein of interest, connected via a linker protein to an anchoring protein native to the spore. This technique offers the added advantage of using the sporulation process itself for protein production, compared to e.g. adsorption based methods of displaying proteins on the spore surface3.
The formed spores can be subdivided into three sections, namely the core, the inner & outer coat and outermost layer, the crust. Each layer incorporates a distinct set of proteins which can be used as anchoring proteins.
Their physical properties compared to dissolved enzymes, leads to them being readily purified by centrifugation and repeated washing steps4.
After having successfully employed spore display to immobilize a photodecarboxylase5 and sucrose phosphorylase, one of our current research goals is to utilize spore display to produce diagnostics enzymes currently in high demand due to the Covid-19 epidemic. We believe that our system can be especially helpful in low ressource environments such as rural Africa, because it allows for an easy, de-skilled production of said enzymes right at the point of care.

References
- Becker, M., Lütz, S. & Rosenthal, K. Environmental Assessment of Enzyme Production and Purification. Molecules 26, Enzyme Microb. Technol. 49, 66–71 (2021)
- Guoyan, Z. et al. Bacillus subtilis Spore Surface Display Technology: A Review of Its Development and Applications. J. Microbiol. Biotechnol. 29, 179–190 (2019)
- Cho, E.-A., Kim, E.-J. & Pan, J.-G. Adsorption immobilization of Escherichia coli phytase on probiotic Bacillus polyfermenticus spores. Enzyme Microb. Technol. 49, 66–71 (2011)
- Tavares, M. B. et al. Bacillus subtilis endospores at high purity and recovery yields: optimization of growth conditions and purification method. Curr. Microbiol. 66, 279–285 (2013)
- Karava, M., Gockel, P., Kabisch, J. & Kabisch, J. Bacillus subtilis spore surface display of photodecarboxylase for the transformation of lipids to hydrocarbons. bioRxiv doi:10.1101/2020.08.30.27382, (2020)
Bruna Schuck
Bruna Schuck1, Vipul Panchal1, Ruth Brenk1
1 Department of Biomedicine, University of Bergen
Antibiotic resistance looms as a serious threat to our well-being, health care systems and, consequently, the global economy as we continue to overuse and misuse antibiotics. Therefore, the discovery and development of new antibiotics are urgently needed. The 2C-methyl-D-erythritol 4-phosphate (MEP) pathway generates precursors of isoprenoids which are natural products essential for many human pathogens. As this pathway is absent in humans, its enzymes are attractive targets for new antibiotics.
In this project, we work with 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR), the second enzyme of the MEP pathway, from Pseudomonas aeruginosa (PaDXR) and Mycobacterium tuberculosis (MtDXR). MtDXR is used mainly as a comparator since almost no data is available for PaDXR and much is already known about MtDXR. A plasmid for each was designed with a 6xHis-tag, Avi-tag, and TEV protease cleavage site. After a successful transformation in E. coli BL21(DE3), MtDXR, and E. coli Rosetta(DE3)pLysS, PaDXR, the proteins were expressed and purified. Subsequently, the process was optimized and the best buffer conditions for each protein were established. The enzymatic activity for PaDXR was confirmed by a DXS-DXR coupled assay.
Next, binding experiments using bio-layer interferometry (BLI) were performed. The conditions were optimized for PaDXR, using MgCl2 as the salt, in order to reproducibly determine the binding of a series of known ligands (NADPH and NADP+, 1-deoxy-D-xylulose 5-phosphate (DXP), fosmidomycin and analogs). So far, only binding for the co-enzymes could be determined (NADPH, KD = 11.9 ± 1.6 µM; NADP+, KD = 42.0 ± 1.4 µM). As the molecular weight of fosmidomycin is close to the detection limit of BLI, the enzymatic assay is currently being used as an orthogonal binding assay. Further, enzymatic and BLI experiments with MtDXR are underway to characterize compound binding to this enzyme. Once the binding affinity of fosmidomycin could be asserted using the coupled assay, a fragment screening of in-house libraries available at the University of Bergen will be carried out.

Flore Kersten
Flore Kersten1,2, Gabriele Cordara2, Stefanie Schmieder3, Markus Künzler3, Ute Krengel2
1 NCMM, University of Oslo, Norway
2 Department of Chemistry, University of Oslo, Norway
3 Institute of Microbiology, Department of Biology, ETH Zürich, Switzerland
CCTX2 is a 89 kDa chimerolectin produced in the vegetative mycelium of the fungus Coprinopsis cinerea (grey shag) and acting as defence protein against fungivorous nematodes [1]. CCTX2 contains four ricin B chain-like lectin domains and a fifth domain, responsible of the cytotoxic activity. The fifth domain displays a “RDQ motif”, a signature sequence found in Poly-ADP-Ribose Polymerases (PARPs). Accordingly, preliminary in vitro experiments suggest a NAD-dependent self-ribosylating activity of CCTX2 [2]. However, the lack of significant sequence conservation with other PARPs outside of the RDQ-motif and the unknown fold of the fifth domain prevent the unambiguous identification of its enzymatic activity, or the full understanding of the reaction mechanism. We have determined the structure of CCTX2 using single particle cryo-EM. Our results shows the first high-resolution structure of CCTX2 as well as complementary data on a cleaved version of the protein, possibly revealing a biologically relevant intermediate. The structure of the fifth domain does not match any of the folds found the in the PDB database and may constitute a completely new family of PARPs.
References
- Plaza, D.F., Schmieder, S.S., Lipzen, A., Lindquist, E., and Künzler, M. (2015). Identification of a novel nematotoxic protein by challenging the model mushroom Coprinopsis cinerea with a fungivorous nematode. G3 (Bethesda) 6, 87-98.
- Schmieder, S.S. (2015). The spatiotemporal regulation and mechanism of action of fungal nematotoxic defense effectors. PhD thesis, doi: 10.3929/ethz-a-010607472.
Greta Daae Sandsdalen
Greta Daae Sandsdalen1, Maryam Imam2, Ole Morten Seternes3 and Hanna-Kirsti Schrøder Leiros1
1 Biomolecular and Structural Chemistry, Department of Chemistry, UiT The Arctic University of Norway
2 The Norwegian College of Fishery Science, UiT The Arctic University of Norway
3 Department of Phamacy, UiT The Arctic University of Norway
The CRISPR-Cas genome editing system has revolutionized molecular biology, providing an array of biotechnological tools for carrying out precision genome modification and regulation in eukaryotic cells. One limitation of the system at present is that available tools are developed from and optimized for mesophilic organisms, which limits their utility in extremophilic organisms. Although thermophilic Cas homologs have been developed and verified for use at high temperatures, less attention has been given to the low-temperature end of the spectrum. This paucity of available psychrophilic genome editing tools is particularly problematic for researchers of cold-blooded eukaryotes such as fish biologists, as both the current enzymes and fish cells lose viability at elevated temperatures.
This project is one of three interdisciplinary components of the UiT Strategic-Funded ‘FISH&CRISPR Innovative strategies to improve salmon health’ which aims to establish a platform for the development of a low-temperature CRISPR-Cas genome editing system optimized for salmonids. In this project, the goal is to discover and develop one or more CRISPR-associated endonucleases for effective and precise genome editing at low temperatures. Candidates were selected by bioinformatics analysis of genomes of psychrophilic bacteria.
Currently we are focusing on optimizing CRISPR activity assays to confirm the cold-active nature of a Cas12a endonuclease in addition to optimization of purification protocols for the Cas9 target enzymes. The goal is that one or more of these Cas enzymes will be characterized, including biophysical properties, enzymatic properties, and structure. The ultimate goal is verification that the Cas/sgRNA complexes are able to perform efficient and accurate gene editing in salmon cell lines at low temperatures.

Katja Stangeland Håheim
Katja S. Håheim1 and Magne O. Sydnes1
1 Faculty of Science and Technology, Department of Chemistry, Bioscience and Environmental Engineering, University of Stavanger
Indoloquinolines are an important class of bioactive compounds whose structural motifs are found in natural products, pharmaceuticals, agrochemicals and drug candidates[1, 2]. The most notable pharmacological properties found in indoloquinolines are antimalarial [3], antiproliferative [4] and antibacterial [5].
Building on strategies from our previous synthetic endeavors to create indoloquinolines [6,7] it became apparent that it would be feasible to synthesize both indolo[2,3-b]quinolines (i.e. neocryptolepines) 2 and indolo[3,2-c]quinolines 3 from a common starting material 1. Neocryptolepines 2 were prepared by N-alkylation of haloquinolines 1 followed by a tandem Suzuki-Miyaura cross-coupling reaction and intramolecular cyclization. To furnish indolo[3,2-c]quinolines 3, a lengthier approach was necessary, the key synthetic strategies being a standard Suzuki-Miyaura cross-coupling reaction followed by diazotization-azidation and finally photochemical cyclization. The viability of the two pathways were evaluated by conducting a large scope and limitations study, which revealed a broad tolerance to both EDGs (e.g. OMe and Me) and EWGs (e.g. CN, CF3 and F), however, nitro-functionalizations were overall poorly tolerated.

References
- Zhu, J.-K.; Gao, J.-M.; Yang, C.-J.; Shang, X.-F.; Zhao, Z.-M.; Lawoe, R. K.; Zhou, R.; Sun, Y.; Yin, X.-D.; Liu, Y.-Q. J. Agric. Food Chem., 2020, 68, 2306-2315.
- Symington, S. B., Zhang, A., Karstens, W., Van Houten, J., Clark, J. M. Pestic. Biochem. Phys., 1999, 65, 181-193.
- Sofowora, A. Medicinal Plants and Traditional Medicine in Africa; John Wiley & Sons: Chichester, UK, 1982; pp. 183-256.
- Lu, C.-M., Chen, Y.-L., Chen, H.-L., Chen, C. A., Lu, P.-J., Yang, C. N., Tzeng, C.-C. Bioorg. Med. Chem., 2010, 18, 1948-1957.
- Paulo, A., Duarte, A., Gomes, E. T. J. Enthnopharmacol., 1994, 44, 127-130.
- Håheim, K. S., Helgeland, I. T. U., Lindbäck, E., Sydnes, M. O. Tetrahedron, 2019, 75, 2924-2957.
- Håheim, K. S., Linbäck, E., Tan, K. N., Albrigtsen, M., Helgeland, I. T. U., Lauga, C., Matringe, T., Kennedy, E. K., Andersen, J. H., Avery, V. M., Sydnes, M. O. Molecules, 2021, 26, 3268-3290.
Lisa Schroer
Lisa Schroer, Erna Davydova and Pål Falnes
University of Oslo
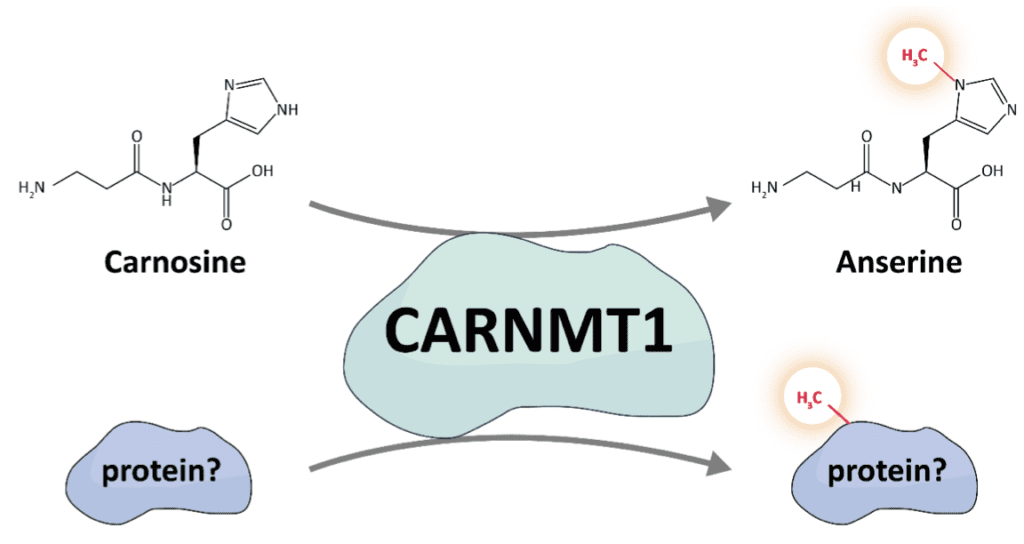
Carnosine methyltransferase 1 (CARNMT1) is the responsible catalyst of the formation of anserine (π-methylation) from carnosine. Already in the original characterization of the MTase Drozak et al. have shown that CARNMT1, is a rather broad specific enzyme, methylating not only carnosine but also L-histidine itself and L-histidine-containing peptides 1 . Furthermore, it has been found that CARNMT1 is a highly conserved MTase, whereas the enzyme generating carnosine, is not. This, in addition to the promiscuous methylation of L-histidine containing peptides in vitro leads to the hypothesis that there are yet undiscovered substrates of CARNMT1, which can potentially include proteins 1,2 .
In this presentation, we followed up on this hypothesis and investigated the potential of CARNMT1 to methylate proteins and longer peptides as well. If you want to know if CARNMT1 indeed has a taste for proteins, come to my presentation and/or poster!
Footnotes:
- Drozak, J. et al. UPF0586 protein C9orf41 homolog is anserine-producing methyltransferase. J. Biol. Chem. (2015). doi:10.1074/jbc.M115.640037
- Kwiatkowski, S., Kiersztan, A. & Drozak, J. Biosynthesis of Carnosine and Related Dipeptides in
Vertebrates. Curr. Protein Pept. Sci. (2018). doi:10.2174/1389203719666180226155657
Typhaine Le Doujet
Utilizing the gut microbiome of Atlantic cod as a platform for the development of cold-active enzymes
Le Doujet Typhaine1, Haugen Peik1
1 UiT – The Arctic University of Norway
The migrating Atlantic cod, a cold-water fish, comprises the world’s largest population of Atlantic cod. It is an interesting subject for microbiota/microbiome studies because of its distinct life cycle, migration pattern, feeding resources, and economic and cultural importance. The cod intestine is of particular interest for microbiome studies because it interacts directly with its environment, and helps in several processes including digestion. During evolution, its microbiota has evolved to exploit structural features and energy resources found in the gut, and over time, genes that benefit the microbes while minimizing negative impacts on the host are enriched. Given this assumption, the gut microbiota of marine fish is expected to contain bacteria with genes highly specialized for breaking down fibrous proteins (i.e., collagen, keratin, muscles), carbohydrates (e.g., chitin) and other macromolecules. Surprisingly, this obvious source of industry-relevant enzymes and small molecules is largely overlooked. Recently, we published the bacterial diversity as well as the functional profile and metaproteome of the gut of migrating Atlantic cod. Here, we describe how those results can be used as a platform for mining of commercially interesting cold-active enzymes (e.g. chitinase, collagenase, trypsin, aminopeptidase P). In addition, we will present Aliivibrio wodanis as a production host for expressing identified cold adapted enzymes in the gut of Atlantic cod.

Yomkippur Perez
Yomkippur Perez1, Erik Agner2 and Magne Olav Sydnes3
1 Polypure AS and University of Stavanger (UiS), 2 Polypure AS, 3 UiS
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive forms of cancer, killing about 95% of patients within 5 years of diagnosis. Despite extensive efforts, traditional therapies such as chemotherapy and radiotherapy have not shown significant improvement in overall survival rates. Surgery is only applicable for those in the early stages of the disease however, most patients have locally advanced or metastatic PDAC at the time of diagnosis. In recent years, immunotherapy-based strategies have shown promising results in treating various types of cancers and managing minimal residual disease after pancreatic resection. Our project is part of a multidisciplinary group that will develop novel immunotherapy approaches for the treatment of PDAC. In this study, a multi-component nanovaccine comprising of tumor antigens, immunomodulators, and imaging contrast agents will be developed and delivered through nanocarriers. Additional surface functionalization with various PEG derivatives will also be done. Finally, the efficacy of the multi-component nanovaccine will be tested in vivo in mice and porcine models.

References:
- O’Kane, G. M.; Ladak, F.; Gallinger, S. Advances in the Management of Pancreatic Ductal Adenocarcinoma. Canadian Medical Association Journal 2021, 193.
- Sturm, N.; Ettrich, T. J.; Perkhofer, L. The Impact of Biomarkers in Pancreatic Ductal Adenocarcinoma on Diagnosis, Surveillance and Therapy. Cancers 2022, 14, 217.
- The illustration was created using BioRender.
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 861190
Zuzanna Justyna Samol
Zuzanna Samol1, Erik Agner2
1 Polypure AS, Norway, Department of Chemistry, Bioscience and Environmental Engineering, University of Stavanger, Norway,
2 Polypure AS, Norway
It is estimated that approximately 10% of the general population suffers from neuropathic chronic pain. However, currently available treatments are often based on centrally-acting drugs with systemic adverse effects with only short-term pain relief.
The main aim of the project is to develop and characterize a multifunctional hydrogel patch for the delivery of analgesics. This material will be based on PEG-PPG-PEG hydrogel matrix with incorporated PEGylated PLGA nanoparticles loaded with capsaicin. The focus will be on the chemical composition and purity of each biomaterial component. This approach could potentially lead to highly reproducible biomaterial with increased therapeutic efficacy and little to no side effects.
The current focus is on developing a suitable hydrogel matrix based on PEG-PPG-PEG triblock copolymers (poloxamers). These copolymers will be characterized with a specific length for each polymer block and strictly defined molecular weight. As a proof-of-concept, a triblock copolymer of PEG15-PPG6-PEG15 has been synthesized and characterized. The future perspectives for this work include developing a library of monodisperse PEG-PPG-PEG triblock copolymers with varying lengths of the blocks and derivatization of the terminal groups with functional moieties.
This work has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Actions grant agreement No 956477.
Maria Wilhelmsen Hoff
Maria Wilhelmsen Hoff1, Lindsey Martinsen2, Sietske S. Grijseels3, Terje Vasskog3, J. Johannes Eksteen2 and Peik Haugen3
1 UiT Norges arktiske universitet, 2 Amicoat, 3 UiT Norges arktiske universitet
Sustainable industrial food production and reduction of waste are recognized by the European Agency (EU) as essential parts of the future bioeconomy. Sustainable production entails the entire process from primary production to processing of a high-quality product, waste reduction and waste management1,2 Several recent EU-funded projects have directly target upcycling of biological waste as important measures. Bioconversion by fermenting microorganisms have been explored as potential methods for increased value of underutilized materials3,4,5. Accumulation and storage of polymers by microorganisms can be exploited to recover nutrients from low value materials, with polyhydroxyalkanoates representing a well-known case4,5,6,7. Traditionally, the properties of these microbial polymers have been studied as candidates for biodegradable plastic. Multiple potential usages are emerging due to their compositional flexibility and biocompatibility. However, there are several challenges, including expensive production methods and variable product quality, which must be overcome before sustainable and economically viable PHA production is a reality4,5,6,7. Some of these challenges could be overcome by use of Arctic marine bacteria. Arctic marine bacteria have been found to produce PHA naturally8, and their growth at lower temperatures could reduce the environmental footprint and production costs9. The resulting biomass could be implemented as part of fish feed. This is highly relevant, as aquaculture have been singled out as a key food production industry to meet the nutritional needs of a growing population. Here, the use of plant-based sources can be reduced and bring the aquaculture industry closer to circular food production1,2,3,4,10.
References:
- European Commission (2020). A farm to Fork Strategy for a fair, healthy and environmentally-friendly food system. EUR-Lex – 52020DC0381 – EN – EUR-Lex (europa.eu)
- European Commission (2018). A sustainable Bioeconomy for Europe: Strengthening the connection between economy, society and the environment. EUR-Lex – 52018DC0673 – EN – EUR-Lex (europa.eu)
- European Commission (2020). Bioeconomy. Bioeconomy | Research and Innovation (europa.eu)
- European Commission (2021). Upcycling innovation to extract value from biowaste. Upcycling innovation to extract value from biowaste | Research and Innovation (europa.eu)
- European commission (2021). Extracting value from waste to deliver high-end products. Available from: Extracting value from waste to deliver high-end products | Research and Innovation (europa.eu)
- Tan et al. 2021, Trends in biotechnology, 39(9):953–963.
- Pérez-Rivero et al. 2019, Biochemical Engineering Journal 150:107283.
- Christensen et al. 2021, Microbial Cell Factories 20:225.
- Söderberg et al. 2019, Microbial Cell Factories 18:197.
- Hua et al. 2019, One Earth, 3:316-329.
Marta Hammerstad
Marta Hammerstad1, Hilde Kristin Andersen1, Ingvild Gudim1, and Hans-Petter Hersleth1
1 Department of Biosciences, University of Oslo, P.O.Box 1066 Blindern, NO-0316 Oslo, Norway
Structural studies show that enzymes have a limited number of unique folds, although structurally related enzymes have evolved to perform a large variety of functions. Thioredoxin reductase-like ferredoxin/flavodoxin NAD(P)+ oxidoreductases (FNRs) typically catalyze the electron transfer between NAD(P)H and the Fe–S cluster of ferredoxin or the FMN group of flavodoxin, however, homologs have been shown to be involved in various redox pathways, catalyzing a wide range of reactions. Bacillus cereus encodes three potential FNRs, clustering in three different phylogenetic groups. Through structural and functional investigations, we have demonstrated three distinct functions for these enzymes. FNR1 (or IruO; iron utilizing oxidoreductase) has been shown to function in the reduction of the heme-degrading enzyme IsdG, as well as hydroxamate-type siderophores, and hence serves an important role in iron acquisition. FNR2 has been established as an endogenous redox partner involved in the activation of the flavodoxin-like protein NrdI, and hence in the activation of class Ib ribonucleotide reductase enzyme. FNR3 (or Bdr; bacillithiol disulfide reductase), on the other hand, reduces the oxidized form of bacillithiol, a low-molecular weight thiol used to buffer the intracellular redox environment and counteract oxidative stress in Firmicute bacteria. The conservation of the structural architecture among these FNRs illustrates how enzymes sharing the same structural organization can catalyze a diverse set of chemical reactions, however, modulated by small structural modifications.















