PhD Poster Abstracts 2023
An abstract booklet in PDF format is available for download here, including all poster and oral presentation abstracts. Only the feedback committees will receive a printed version of this booklet at the conference. Feel free to print your own and bring it along. Digital version of the poster abstracts are below. Digital versions of the oral presentation abstracts can be found here.
BioCat Annual Conference 2023 Abstract Book – PDF
Camilla F. Angeltveit
Camilla Fløien Angeltveit1, Tina Jeoh2, Svein Jarle Horn1
1 Faculty of Chemistry, Biotechnology, and Food Science, Norwegian University of Life Sciences (NMBU), Ås, Norway,
2 Biological and Agricultural Engineering, University of California, Davis, United States
Cellobiohydrolases are key enzymes for cellulose degradation, but their activity on crystalline cellulose is limited due to their need for single cellulose chain ends to perform hydrolysis. Thus, cellobiohydrolases rely on other enzymes such as Lytic polysaccharide monooxygenases (LPMOs), which act directly on the surface of the crystalline substrate (Horn et al. 2012), although exactly how LPMOs promote cellulase activity remains to be elucidated.
In this study, we investigated the synergistic relationship between LPMOs and cellulases by determining how LPMO pretreatment of crystalline cellulose affected the productive binding capacity (i.e., sites where the enzyme can complex and hydrolyze the substrate) of a reducing-end cellobiohydrolase from Trichoderma longibrachiatum (TlCel7A) on cellulose. This was done by using an electrochemical three-electrode system with an in-house made cellobiose dehydrogenase biosensor enabling real-time measurement of β-cellobiose. Assuming that cellobiose release during the burst phase of the reaction is a direct indication of productively bound enzymes, we evaluated the effect of LPMOs on the substrate by calculating the productive binding capacity of TlCel7A. A significant increase in the productive binding capacity of TlCel7A on cellulose was detected between 5 and 24 hours of LPMO, even though all LPMO reactions were inactivated already at the first time point. Hence, the increased productive binding capacity between the two time points cannot be explained by further introduction of oxidized cuts by the LPMOs. This delayed LPMO effect suggests that LPMO cleavage of cellulose is followed by a gradual decrystallization of the substrate that makes the substrate more accessible for the cellulases (Koskela et al. 2023; Uchiyama et al. 2022).
Regardless of the LPMO regioselectivity (i.e., C1 or C4 activity), it was found that 24 hours of LPMO pretreatment of cellulose resulted in an increased productive binding capacity of TlCel7A on cellulose. This study suggests, in accordance with other recent studies (Koskela et al. 2023; Uchiyama et al. 2022), that the cellulose chain cleavage by LPMOs in itself does not lead to the improved binding capacity for cellulases, but ratherthedecrystallization of the cellulose following the oxidative cleavage.
References:
(1) Horn, S. J., Vaaje-Kolstad, G., Westereng, B. & Eijsink, V. (2012). Novel enzymes for the degradation of cellulose. Biotechnology for biofuels, 5 (1): 45.
(2) Koskela, S., Wang, S., Li, L., Zha, L., Berglund, L. A. & Zhou, Q. (2023). An Oxidative Enzyme Boosting Mechanical and Optical Performance of Densified Wood Films. Small: 2205056.
(3) Uchiyama, T., Uchihashi, T., Ishida, T., Nakamura, A., Vermaas, J. V., Crowley, M. F., Samejima, M., Beckham, G. T. & Igarashi, K. (2022). Lytic polysaccharide monooxygenase increases cellobiohydrolases activity by promoting decrystallization of cellulose surface. Science Advances, 8 (51): eade5155.
Antonia Areali
Areali Antonia1
1 NTNU
Secretion systems in bacteria are of high scientific importance, as various molecules can be secreted from bacteria to respond in different processes. Many factors can affect bacterial secretion systems and the export of proteins, therefore understanding of those factors is the key for a proper biotechnological exploitation of microbial secretion systems. Currently, several bacteria workhorses like Escherichia coli have been extensively studied and engineered to produce high value proteins. The purpose of this research is to study, engineer, optimize and test for secretion limitations regarding protein size in the flagellar secretion systems T3SS which is one of the most important secretion systems in bacteria and has shown a remarkably fast exportation of proteins to the medium. As proof-of-concept, within this project we will establish biotechnological production of high value proteins as, for instance, protein-based fish vaccines at different production scales.
Mina Bathen
Mina Bathen1, Åsmund Haatveit1, Anders Vik1 and Trond Vidar Hansen1
1 Department of Pharmacy, Section for Pharmaceutical Chemistry, University of Oslo, PO Box 1068 Blindern, N-0316 Oslo, Norway
Epoxy fatty acids (EpFAs) are biosynthesised from dietary polyunsaturated fatty acids (PUFAs) by the enzyme cytochrome P450 (CYP450).1 The EpFAs are lipid mediators with potent anti-inflammatory2 and pain relief effects.3 The most important degradation pathway for the EpFAs is hydrolysis by the enzyme soluble epoxide hydrolase (sEH), which gives the corresponding dihydroxy fatty acids.
The PUFA-product 4S,5S-dihydroxy docosapentaenoic acid (1) could be the product of such an enzymatic process. This PUFA has been reported from various human samples4 and has not yet been the target of stereoselective total synthesis. This poster presents our current work on a stereoselective semi-synthesis of 1 from docosahexaenoic acid (DHA).

References:
(1) Morisseau, C.; Hammock, B. D. Impact of Soluble Epoxide Hydrolase and Epoxyeicosanoids on Human Health. Annu. Rev. Pharmacol. Toxicol. 2013, 53 (1), 37-58.
(2) Wagner, K. M.; McReynolds, C. B.; Schmidt, W. K.; Hammock, B. D. Soluble epoxide hydrolase as a therapeutic target for pain, inflammatory and neurodegenerative diseases. Pharmacol. Ther. 2017, 180, 62-76.
(3) Wagner, K.; Inceoglu, B.; Hammock, B. D. Soluble epoxide hydrolase inhibition, epoxygenated fatty acids and nociception. Prostaglandins Other Lipid Mediat. 2011, 96 (1-4), 76-83.
(4) (a) Lundström, S. L.; Yang, J.; Brannan, J. D.; Haeggström, J. Z.; Hammock, B. D.; Nair, P.; O’Byrne, P.; Dahlén, S.-E.; Wheelock, C. E. Lipid mediator serum profiles in asthmatics significantly shift following dietary supplementation with omega-3 fatty acids. Mol. Nutr. Food Res. 2013, 57 (8), 1378-1389. (b) Schuchardt, J. P.; Schneider, I.; Willenberg, I.; Yang, J.; Hammock, B. D.; Hahn, A.; Schebb, N. H. Increase of EPA-derived hydroxy, epoxy and dihydroxy fatty acid levels in human plasma after a single dose of long-chain omega-3 PUFA. Prostaglandins Other Lipid Mediat. 2014, 109-111, 23-31.
Marcus de Bourg
Marcus de Bourg1, Rawand S. Mohammad1, Abhishek Mishra2, Ashraf El-Meanawy4, Jawad Bin Belayet4, Bruce D. Hammock3, John D. Imig2 and Anders Vik1
1 Section for Pharmaceutical Chemistry, Department of Pharmacy, University of Oslo, Norway.
2 Department of Pharmaceutical Sciences, University of Arkansas for Medical Sciences, Little Rock, AR, USA.
3 Department of Entomology and Nematology, University of California at Davis, Davis, CA, USA .
4 Department of Medicine, Medical College of Wisconsin, Milwaukee, WI, USA
Epoxy fatty acids (EpFAs) are linked to several bioactions in the body. Some of these include pain-relief, anti-inflammatory effect.However, several biological pathways quickly degrade the EpFAs, with the dominating mechanism being epoxide hydrolysis by the enzyme soluble epoxide hydrolase (sEH). Increasing EpFA concentrations through inhibition of the sEH has shown to reduce the severity of a variety of disease states in laboratory settings.1 In particular, 8,9-epoxyeicosatrienoic acid (8,9-EET) has proved effective at protecting the glomerular filtration barrier.2
The focus of this project is the synthesis and biological evaluation of potential EpFA mimics that are stable towards sEH. A total of 19 stable potential mimics of 8,9-EET were synthesized. These candidates were tested for their ability to protect glomerular mesangial cells from Sorafenib induced nephrotoxicity.

References
(1) Morisseau, C.; Hammock, B. D. Annual Review of Pharmacology and Toxicology 2013, 53, 37.
(2) Sharma, M.; McCarthy, E. T.; Reddy, D. S.; Patel, P. K.; Savin, V. J.; Medhora, M. & Falck, J. R. Prostaglandins & other lipid mediators, 2009, 89, 43-51
Mateu Montserrat Canals
Mateu Montserrat-Canals1,2, Kilian Schnelle3, Vilde Leipart4, Øyvind Halskau5, Gro Amdam4,6, Arne Moeller3, Harmut Luecke7 and Eva Cunha
1 Department of Chemistry, University of Oslo, 0315 Oslo, Norway; Norwegian Centre for Molecular
2 Medicine, University of Oslo, 0318 Oslo, Norway; Department of Structural Biology, Osnabrück
3 University, 49076 Osnabrück, Germany;
4 Faculty of Environmental Sciences and Natural Resource Management, Norwegian University of Life Sciences, Aas, Norway; Department of Biological Sciences, University of Bergen, Norway
5 School of Life Sciences, Arizona State University, Tempe, AZ, United State
6 NOVA School of Science and Technology, Lisbon, Portugal
Vitellogenin (Vg) is the main yolk precursor protein in almost all egg laying animals. In addition, along its evolutionary history, Vg has developed a range of new functions in different taxa. In the honey bee, Vg has functions related to immunity, antioxidant protection, social behaviour and longevity. However, the molecular mechanisms underlying Vg functionalities are still poorly understood. Here, we report our cryo-EM studies on Vg purified directly from the haemolymph of honey bees.
Carina Dietz
Carina Dietz1, Johannes Kabisch1
1 Department of Biotechnology and Food Science, NTNU
Bacillus subtilis (B. subtilis) is a Gram-positive soil bacterium that, in its vegetative state, is an important workhorse in the biotechnology industry for the production of enzymes, food additives and vitamins etc. It further contains the ability to respond to environmental stress by forming a biofilm or sporulating (Su et al., 2020). Large-scale production of these spores has already been developed for use in agriculture as plant pathogen antagonists and plant growth promoters (personal communication, Tobias May, BASF). The spores are also being developed for use in self-healing concrete, drug delivery and as protein immobilisation platforms (Zhang et al., 2020). The latter uses fusion protein technology in order to anchor the target protein on to the spore surface using an anchoring protein which is naturally present on the spore surface. This process allows for rapid immobilisation of the protein and easy purification, as only a simple centrifugation process is required (Karava et al., 2021). However, this project aims to extend the use of spores as a platform beyond protein display by developing a novel and programmable hybrid biomaterial. This will require the development of a non-germinating spore chassis to potentially enable the use of B. subtilis spores outside of a laboratory environment. Currently developed mutant strains can reduce the germination rate to <0.002%, but a 100% tight containment system has not yet been developed (Karava, 2021). We will also use the principle of spore display to present fibre-forming proteins on the spore surface to create a hybrid material consisting of inert spores and the displayed proteins. This will allow the development of a process that overcomes two of the major challenges in developing a biotechnological process: a) scale-up, since the large-scale production of bacterial spores has already been developed, and b) downstream processing, since the use of spore display does not require extensive purification steps but rather a centrifugation process (Karava, 2021).
References:
(1) Karava, M. (2021). Development of a platform for immobilization of proteins based on Bacillus subtilis spores [Ph.D. Thesis, Technische Universität]. https://doi.org/10.26083/tuprints-00019729
(2) Karava, M., Gockel, P., & Kabisch, J. (2021). Bacillus subtilis spore surface display of photodecarboxylase for the transformation of lipids to hydrocarbons. Sustainable Energy & Fuels, 5(6), 1727–1733. https://doi.org/10.1039/D0SE01404D
(3) Su, Y., Liu, C., Fang, H., & Zhang, D. (2020). Bacillus subtilis: A universal cell factory for industry, agriculture, biomaterials and medicine. Microbial Cell Factories, 19(1), 173. https://doi.org/10.1186/s12934-020-01436-8
(4) Zhang, X., Al-Dossary, A., Hussain, M., Setlow, P., & Li, J. (2020). Applications of Bacillus subtilis Spores in Biotechnology and Advanced Materials. Applied and Environmental Microbiology, 86(17), e01096-20. https://doi.org/10.1128/AEM.01096-20
Karina Ervik
Karina Ervik1, Amalie F. Reinertsen1 and Trond V. Hansen1
1 Section for Pharmaceutical Chemistry, Department of Pharmacy, University of Oslo, P.O. Box 1068, 0316 Oslo, Norway
Inflammation is divided into acute and chronic inflammation. Chronic inflammation occurs over a longer period of time, and remains unresolved.1 Uncontrolled chronic inflammation can result in the progress of several diseases such as cancer, rheumatoid arthritis and neurological disorders. Recently, it was discovered that the resolution phase of inflammation and the return to homeostasis are regulated by active and strictly controlled biosynthesis of several novel families of oxygenated polyunsaturated fatty acid products.2 These endogenously formed compounds have been coined specialized pro-resolving mediators (SPMs) and display numerous novel and interesting bioactivities.3
The SPM RvD5n-3 DPA (1)4 is an excellent biotemplate for the development of new, small molecular anti-inflammatory drugs and immunoresolvents.
In this poster, the first total synthesis of 1 is presented, based on the retrosynthetic outline presented below.

References:
(1) Tabas, I., Glass, C. K., Science, 2013, 339, 166-172.
(2) Serhan, C. N., Petasis, N. A., Chem. Rev. 2011, 111, 5922-5943.
(3) Serhan, C. N., Nature, 2014, 510, 92-101.
(4) Dalli, J., Colas, R. A., Serhan, C. N., Sci. Rep. 2013, 3, 1940.
Philipp Garbers
Philipp Garbers1, Sara M. Gaber2, Hans A. Brandal1, Aksel V. Skeie1, Svein H. Knutsen2 Catrin Tyl1, and Bjørge Westereng1
1 Norwegian University of Life Science (NMBU), Ås, Norway
2 Nofima AS, Ås, Norway
Pulses like peas or faba beans are interesting in the shift towards a more sustainable food ecosystem and healthier diets due to the plants ability to fixate nitrogen as well as their constituents with favourable nutritional effects, most notably protein and dietary fibre. However, they also contain constituents of potential concern such as fermentable oligo-, di- and monosaccharides and polyols (FODMAPs), most significantly the raffinose family oligosaccharides (RFOs).
The aim of this work is therefore to develop a sustainable and industrially feasible process for the extraction of RFOs from protein concentrates and residuals derived from pulses. At the same time, the aim is to also utilize the extracted oligosaccharides to produce new and potentially value-added food products or ingredients. To extract the oligosaccharides from the milled plant material, a water-based pilot-scale process was designed that utilizes solubilization, separation, and filtration technology to produce a concentrated RFO extract as well as a protein enriched fraction. The fractions were freeze-dried and characterized. The RFO extract was furthermore used as carbon source in fermentation studies with food-grade microorganisms, for example as a possibility to enhance the growth of lactic acid bacteria in modern sour beer production as previously demonstrated with wood derived oligosaccharides. Furthermore, the potential enzymatic modifications of RFOs are researched through recombinant enzyme production and activity screenings.
The developed process allowed for kilogram-scale extraction of high purity RFOs from pea protein concentrates. The residual material after extraction had on the other hand increased protein and decreased oligosaccharide content in comparison to the starting material. The freeze-dried extract was shown to be a suitable carbon source for a variety of food-grade microorganisms and was successfully applied as an additional carbon source for these organisms in the production of sour beer. In a first step towards enzymatic modifications, activity of pure enzymes could be shown on the obtained RFO extract as well.
References:
(1) Green Technology for Plant Based Food, Nofima AS, 2023 – nofima.com/projects/gree-technology-for-plant-based-food/
(2) Nyyssölä et al., 2020 – Reduction of FODMAP content by bioprocessing –https://doi.org/10.1016/j.tifs.2020.03.004
(3) Dysvik et al., 2019 – Secondary Lactic Acid Bacteria Fermentation with Wood-Derived Xylooligosaccharides as a Tool to Expedite Sour Beer Production – https://doi.org/10.1021/acs.jafc.9b05459
Hemanga Gogoi
Hemanga Gogoi1, Dario Segura-Peña1 and Nikolina Sekulic1*
1 Norwegian Centre for Molecular Medicine (NCMM) and Department of Chemistry, University of Oslo
*All correspondences are to be addressed to nikolina.sekulic@ncmm.uio.no
Aurora B is a serine/threonine protein kinase. Its enzymatic activity is required for accuratechromosome segregation, spindle midzone stability, and cell division. Together withINCENP, Survivin, and Borealin, Aurora B constitutes the chromosome passenger complex (CPC). Aurora B is up-regulated and/or overexpressed in many cancer types, which is always associated with a poor prognosis. Due to its essential regulatory role in cell division, Aurora B has been identified as an attractive target for cancer chemotherapy, and several inhibitors are already in clinical trials. Temperature-sensitive mutants studies have shown that the N terminus of INCENP i.e. neighbouring the Aurora B interacting region has a role in the enzymatic activity. Here, we propose to elucidate the biochemical basis of the temperaturesensitive mutants and a potential role of a previously uncharacterised region of INCENP in the protein kinase activity of Aurora B kinase.
Mina Gravdahl
Mina Gravdahl1, Bjørn E. Christensen1 and Finn L. Aachmann1
1 Norwegian Biopolymer Laboratory (NOBIPOL), Department of Biotechnology and Food Science, NTNU Norwegian Univeristy of Science and Technology, Trondheim, Norway
Alginate is a family of polysaccharides containing β-D-mannuronic acid (M) and α-L-guluronic acid (G) in varying ratio and is the main structural component in brown algae. Alginates are polyelectrolytes, and therefore, inherently pH responsive. Furthermore, oligoG alginate can interact strongly with certain divalent cations, e.g., and . OligoG alginate could therefore be attractive to incorporate in materials to obtain molecules with new or improved properties. Terminal substitution of polysaccharides, which is conjugation to the reducing ends or the non-reducing ends, give rise to architectures with a large variation in physiochemical properties. Compared to lateral conjugation, terminal conjugation preserves the intrinsic properties of polysaccharides to a greater extent.
In this study, pure guluronate oligomers ( ) were prepared by acid precipitation of guluronate rich alginates. Narrow fractions of oligoG were obtained by size-exclusion chromatography. End-functionalized oligoG were prepared by conjugating aminooxy- – at the reducing end of oligoG by reductive amination. The reaction kinetics were studied using NMR.
DBCO reagents is a class of click chemistry labeling reagents which can react with azide-tagged molecules via copper-free click chemistry. The prepared – – conjugates can therefore be conjugated to a second molecule by strain-promoted azide-alkyne click chemistry (Figure 1).
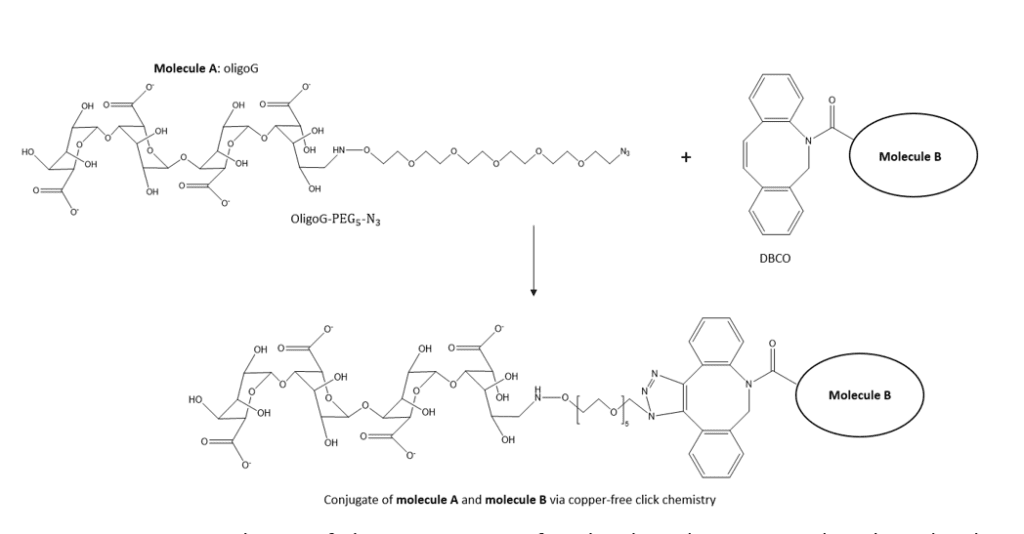
Figure 1: Reaction scheme of the conjugation of molecule A ( – – ) with molecule B via copper-free click chemistry.
Maria Wilhelmsen Hoff
Maria Wilhelmsen Hoff1, Lindsey Martinsen2, Sietske S. Grijseels3, Terje Vasskog1,J. Johannes Eksteen2 and Peik Haugen1
1 UiT Norges arktiske universitet,
2 Amicoat.
Sustainable industrial food production and reduction of waste are recognized by the European Agency (EU) as essential parts of the future bioeconomy. Sustainable production entails the entire process from primary production to processing of a high-quality product, waste reduction and waste management1,2. Several recent EU-funded projects have directly target upcycling of biological waste as important measures. Bioconversion by fermenting microorganisms have been explored as potential methods for increased value of underutilized materials3,4,5. Accumulation and storage of polymers by microorganisms can be exploited to recover nutrients from low value materials, with polyhydroxyalkanoates representing a well-known case4,5,6,7. Traditionally, the properties of these microbial polymers have been studied as candidates for biodegradable plastic. Multiple potential usages are emerging due to their compositional flexibility and biocompatibility. However, there are several challenges, including expensive production methods and variable product quality, which must be overcome before sustainable and economically viable PHA production is a reality 4,5,6,7. Some of these challenges could be overcome by use of Arctic marine bacteria. Arctic marine bacteria have been found to produce PHA naturally8, and their growth at lower temperatures could reduce the environmental footprint and production costs9. The resulting biomass could be implemented as part of fish feed. This is highly relevant, as aquaculture have been singled out as a key food production industry to meet the nutritional needs of a growing population. Here, the use of plant-based sources can be reduced and bring the aquaculture industry closer to circular food production1,2,3,4,10.
References:
(1) European Commission (2020). A farm to Fork Strategy for a fair, healthy and environmentally-friendly food system. EUR-Lex – 52020DC0381 – EN – EUR-Lex (europa.eu)
(2) European Commission (2018). A sustainable Bioeconomy for Europe: Strengthening the connection between economy, society and the environment. EUR-Lex – 52018DC0673 – EN – EUR-Lex (europa.eu)
(3) European Commission (2020). Bioeconomy. Bioeconomy | Research and Innovation (europa.eu)
(4) European Commission (2021). Upcycling innovation to extract value from biowaste. Upcycling innovation to extract value from biowaste | Research and Innovation (europa.eu)
(5) European commission (2021). Extracting value from waste to deliver high-end products. Available from: Extracting value from waste to deliver high-end products | Research and Innovation (europa.eu)
(6) Tan et al. 2021, Trends in biotechnology, 39(9):953–963.
(7) Pérez-Rivero et al. 2019, Biochemical Engineering Journal 150:107283.
(8) Christensen et al. 2021, Microbial Cell Factories 20:225.
(9) Söderberg et al. 2019, Microbial Cell Factories 18:197.
(10) Hua et al. 2019, One Earth, 3:316-32
Selina Cannon Homaei
Selina Cannon Homaei1, Elaheh Mahootchi1, Jarl Underhaug2, and Jan Haavik1,3
1Department of Biomedicine, University of Bergen, Norway,
2Department of Chemistry, University of Bergen, Norway,
3Division of Psychiatry, Haukeland University Hospital, Bergen, Norway
We generated the first animal model for the enzyme Glutamate Decarboxylase Like 1 (GADL1)1. Metabolomic studies reveal that GADL1 decarboxylates aspartate to b-alanine, consequently altering metabolite levels of b-alanine and carnosine peptides in a tissue-specific manner1,2. β-alanine is a precursor for the dipeptide carnosine (b-alanyl-L-histidine) and related histidine-containing dipeptides (HCDs). It is well-known that excitatory tissues are highly abundant in carnosine, specifically the olfactory bulb and skeletal muscle tissue3,4. Carnosine harbors several biological functions, including antioxidation, neurotransmission, metal chelating and pH buffering4. Hence, it is an attractive target for exercise supplements and drug therapy targeting diseases related to oxidative stress, e.g., cancers and neurodegenerative disorders. Although several lines of evidence show that endogenous carnosine levels are age- and sex-dependent3,4, the extent of functional differences is still unclear.
The established GADL1 mouse model was the first tool to investigate the effects of carnosine depletion without giving large pharmacological doses of β-alanine or carnosine1. Our research shows that reduced carnosine levels lead to altered levels of antioxidant enzymes and sex-specific changes in energy and lipid metabolism1. In this PhD project, we aim to further investigate the functional role of GADL1, and carnosine peptides, through behavioral studies, multi-omics and biochemical assays for age-related markers.
References:
(1) Mahootchi, E., et al. (2020). “GADL1 is a multifunctional decarboxylase with tissue-specific roles in beta-alanine and carnosine production.” Sci Adv 6(29): eabb3713.
(2) Winge, I., et al. (2015). “Mammalian CSAD and GADL1 have distinct biochemical properties and patterns of brain expression.” Neurochem Int 90: 173-184.
(3) Van der Stede, T., et al. (2023) “Extensive profiling of histidine-containing dipeptides reveals species- and tissue-specific distribution and metabolism in mice, rats and humans.” BioRxiv Preprint.
(4) Boldyrev, A. A., et al. (2013). “Physiology and pathophysiology of carnosine.” Physiol Rev 93(4): 1803-1845.
Robin Jeske
Robin Jeske1
1 Arctic University of Norway, Department of Chemistry, Norstruct
Peptide therapeutics have gained significant attention in recent years due to their potential as effective drugs for treating a variety of diseases caused by druggable targets[1].
Recent advances in peptide design and computational methods have improved our ability to model and manipulate peptide-mediated interactions[2]. Combining binding site prediction with peptide docking has been shown to be effective in modeling peptide-protein interactions[3].
To take advantage of this advancement to design novel peptides with therapeutic application, understanding of the binding pathway is important.
Flexible protein segments and bound peptides or ligands can populate several microstates in thermodynamic equilibrium, indicating intermediate flexibility. A peptide, for example, can significantly populate multiple microstates in thermodynamic equilibrium[4].
Induced fit cannot generate the arbitrary conformational states required for the biological function of many enzymes. Instead conformational selection accesses various conformational states, and each binding event takes place via conformational selection coupled to a population shift, which allosterically guides the next binding event[1]. Therefore, binding is viewed as a game of induced fit and conformational selection, with one partner’s conformation serving as an environment or set of preconditions for the other partner.

References:
(1) K. Fosgerau and T. Hoffmann, “Peptide therapeutics: current status and future directions,” Drug Discovery Today, vol. 20, pp. 122–128, jan 2015.
(2) G. Weng, J. Gao, Z. Wang, E. Wang, X. Hu, X. Yao, D. Cao, and T. Hou, “Comprehensive evaluation of fourteen docking programs on protein–peptide complexes,” vol. 16, pp. 3959–3969, apr 2020.
(3) O. Schueler-Furman and N. London, eds., Modeling Peptide-Protein Interactions. Springer New York, 2017.
(4) H. Meirovitch, S. Cheluvaraja, and R. White, “Methods for calculating the entropy and free energy and their application to problems involving protein flexibility and ligand binding,” Current Protein & Peptide Science, vol. 10, pp. 229–243, jun 2009.
(5) R. Nussinov, B. Ma, and C.-J. Tsai, “Multiple conformational selection and induced fit events take place in allosteric propagation,” Biophysical Chemistry, vol. 186, pp. 22–30, feb 2014.
Flore Kersten
Flore Kersten1,2, Gabriele Cordara2, Stefanie Schmieder3, Markus Künzler3, Ute Krengel2
1 NCMM, University of Oslo, Norway
2 Department of Chemistry, University of Oslo, Norway
3 Institute of Microbiology, Department of Biology, ETH Zürich, Switzerland
CCTX2 is an 89 kDa chimerolectin expressed in the vegetative mycelium of the fungus Coprinopsis cinerea (grey shag) upon challenge with the nematode Aphelenchus avenae [1]. The protein acts as defense protein against fungivorous nematodes. CCTX2 contains four ricin B chain-like domains, two of which show lectin activity and selective binding of LacdiNAc-containing glycoepitopes. While the ricin B domains are required for cell entry, the nematotoxic activity is associated to a fifth domain, with no known orthologues outside the realm of fungi. The domain displays an “RDQ motif”, a signature sequence found in Poly-ADP-Ribose Polymerases (PARPs) [2]. However, the lack of significant sequence conservation with other PARPs outside of the RDQ-motif and the unknown fold of the fifth domain have so far prevented the unambiguous identification of its possible enzymatic activity, and the full understanding of the toxicity mechanism. Here we present the first high-resolution cryo-EM structure of CCTX2, showing that the structure of the fifth domain does not match any of the folds found the in the PDB database.
References:
(1) Plaza, D.F., Schmieder, S.S., Lipzen, A., Lindquist, E., and Künzler, M. (2015). Identification of a novel nematotoxic protein by challenging the model mushroom Coprinopsis cinerea with a fungivorous nematode. G3 (Bethesda) 6, 87-98.
(2) Schmieder, S.S. (2015). The spatiotemporal regulation and mechanism of action of fungal nematotoxic defense effectors. PhD thesis, doi: 10.3929/ethz-a-010607472.
Oda Caspara Krokengen
Oda Caspara Krokengen*, Arne Raasakka*, Petri Kursula*,**
*Department of Biomedicine, University of Bergen, Bergen, Norway
** Faculty of Biochemistry and Molecular Medicine & Biocenter Oulu, University of Oulu, Oulu, Finland
The myelin sheath enables saltatory conduction of nerve impulses and are crucial for normal nerve function. Securing the tight compaction of the lipid bilayers are ensured by specific myelin protein where some are intrinsically disordered proteins (IDPs) or have intrinsically disordered regions (IDR). IDP(R)s are known to not fold into one single stable structure, but rather into various conformations depending on molecular interactions and amino acid composition. Around 30% of the humane proteome is thought to be comprised of IDRs and are enriched in proteins associated with neurological diseases. Two key membrane binding proteins within the myelin sheath are the myelin basic protein (MBP) and myelin protein zero (P0), the former being an IDP while the latter have an intrinsically disordered region – the cytoplasmic tail of P0 (P0ct). AlphaFold2 and RoseTTAfold revolutionised structural biology with the artificial intelligence learning-based algorithms of protein structure prediction [1, 2]. Using AlphaFold2 for IDPs have been actively discussed in recent years since it is generally believed that AlphaFold2 generates models with low confidence for these proteins.
With the ongoing debate on the use of Alphafold for IDP(R)s, we investigated the structure of murine MBP and human P0ct using AlphaFold predictions in relation to earlier published experimental data from small angle X-ray diffraction and circular dichroism [3, 4], and theoretical hydrophobic cluster analysis.
The results revealed that the predicted models have a-helical segments that are compatible with membrane-binding sites on both proteins positioned at known foldable segments [5]. For both proteins, the AlphaFold2 models suggests more compact structure than experimentally found in solution and may have predicted membrane-bound conformations of MBP and P0ct. Furthermore, the helical segments displayed by AlphaFold2 contain an increased number of order-promoting residues which is also supported by hydrophobic cluster analysis indicating high propensity for the folded segments to gain secondary structure upon lipid membrane binding [5].
References:
(1) Jumper, J., et al., Highly accurate protein structure prediction with AlphaFold. Nature, 2021. 596(7873): p. 583-589.
(2) Baek, M., et al., Accurate prediction of protein structures and interactions using a three-track neural network. Science, 2021. 373(6557): p. 871-876.
(3) Raasakka, A., et al., Membrane Association Landscape of Myelin Basic Protein Portrays Formation of the Myelin Major Dense Line. Scientific Reports, 2017. 7(1): p. 4974.
(4) Raasakka, A., et al., Molecular structure and function of myelin protein P0 in membrane stacking. Scientific Reports, 2019. 9(1): p. 642.
(5) Krokengen, O.C., A. Raasakka, and P. Kursula, The intrinsically disordered protein glue of the myelin major dense line: Linking AlphaFold2 predictions to experimental data. Biochemistry and Biophysics Reports, 2023. 34: p. 101474.
Davide Luciano
Davide Luciano, Jochen Schmid*, Gaston Courtade**
Department of Biotechnology and Food Science, NTNU Norwegian University of Science and Technology, Trondheim, Norway.
*Institute of Molecular Microbiology and Biotechnology, University of Münster, Münster, Germany.
** Department of Biotechnology and Food Science, NTNU Norwegian University of Science and Technology, Trondheim, Norway.
Exopolysaccharides are a diverse class of molecules with a wide range of industrial applications. A major contributor to this class of polymers is xanthan gum, a heteropolysaccharide synthesised by the Gram-negative bacterium Xanthomonas campestris. It is useful in a variety of fields, such as the food, materials and pharmaceutical industries, due to its versatile mechanical properties. Modifications of the polysaccharide are being studied to modulate its properties for more specific applications. Here we present a computational approach to the identification of key residues in the active site of three glycosyltransferases involved in xanthan biosynthesis and their conformational properties. Our goal is the engineering of these three enzymes so that they can incorporate non-natural units into the polysaccharide and tailor its mechanical properties. To shed light on the underlying molecular interactions governing xanthan biosynthesis, we present preliminary in silico findings on the dynamic behaviour of one of these enzymes and its potential substrate binding site residues.
References:
(1) Jochen Schmid, Volker Sieber, and Bernd Rehm. “Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies”. In: Frontiers in Microbiology 6 (2015). issn: 1664-302X. Doi:10.3389/fmicb.2015.00496.
(2) S.R. Salinas et al. “Binding of the substrate UDP-glucuronic acid induces conformational changes in the xanthan gum glucuronosyltransferase”. In: Protein Engineering, Design and Selection 29.6 (2016), pp. 197–207. Doi:10.1093/protein/gzw007.
Joseph Lumba
Joseph P. Lumba1
1 NTNU
Research on health-care monitoring via synthetic materials became widespread due to recent health and pandemic issues. Among these materials, organic-based biosensors find great popularity due to its flexibility in design and ease of production. Organic biosensors are synthetic compounds that can switch between being a good conductor to good insulator, depending on the dopants added or electric field the material is exposed to.1 This field continues to flourish due to various reasons: the abundance of moieties, monomers, and components that can form novel materials, the relative ease of synthesis and upscale, its extensive potential for property modification via molecular level engineering, and the simplicity of device assembly without requirement for extreme conditions, among others. This project will specifically focus on polymer-based transistors as polymers offer even more flexibility in structure and property design, e.g., experiments with different monomers, copolymerization, attachment of one or more groups as side chains, etc. Research on polymeric semiconductors continues to improve the properties and electron mobilities, however, studies on n-type polymers has significantly fallen behind. This is due to several chemical instabilities which are not as prevalent in p-type counterparts. More specifically, this project aims to alleviate the predominant instabilities in n-type polymers by molecular design, selection of best-suited moieties, tuning of material properties, etc. By the end, the aim is to integrate the synthesized materials with high performing p-type polymers to produce fully polymer-based biosensors.
References:
(1) Griggs, S.; Marks, A.; Bristow, H.; McCulloch, I. N-Type Organic Semiconducting Polymers: Stability Limitations, Design Considerations and Applications. J. Mater. Chem. C 2021, 9 (26), 8099–8128. https://doi.org/10.1039/D1TC02048J.
Natalia Mojica Cortes
Natalia Mojica1, Gabriele Cordara1, Ken Teter2, Ute Krengel1
1 Department of Chemistry, University of Oslo,
2 Burnett School of Biomedical Sciences, University of Central Florida
Vibrio cholerae and enterotoxigenic Escherichia coli (ETEC) are two bacterial pathogens responsible for millions of diarrhea cases each year[1]. These pathogens release two similar AB5 toxins that are directly responsible for the severe diarrhea: the cholera toxin (CT) and the heat-labile enterotoxin (LT), respectively. They consist of a catalytically active A subunit anchored to five copies of a cell-binding B-subunit. After cell uptake, the toxins are transported to the endoplasmic reticulum (ER), where the A subunit dissociates from the rest of the toxin, unfolds, and is finally exported to the cytosol with the help of several host chaperones[2],[3]. These steps are essential for cholera and ETEC intoxication, however, they are still poorly understood at the structural level.
This project aims to investigate the interactions mediating toxin activation by host chaperones. This work involves production of the recombinant target proteins, binding and interaction studies, and structural characterization of the protein complexes. We plan to use a combination of structural biology techniques including cryo-EM, X-ray crystallography, and molecular dynamics simulations. We expect that this work provides structural details about a key molecular mechanism of pathogenesis in bacteria, which can serve as a starting point for developing new treatment strategies against diarrheal diseases.
References:
(1) World Health Organization. Cholera. (accesssed Apr 27, 2023) https://www.who.int/news-room/fact-sheets/detail/cholera (2022).
(2) Burress, H., Kellner, A., Guyette, J., Tatulian, S. A. & Teter, K. HSC70 and HSP90 chaperones perform complementary roles in translocation of the cholera toxin A1 subunit from the endoplasmic reticulum to the cytosol. J. Biol. Chem. 294, 12122–12131 (2019).
(3) Teter, K. Toxin instability and its role in toxin translocation from the endoplasmic reticulum to the cytosol. Biomolecules 3, 997–1029 (2013).
Liza Nguyen
Liza Nguyen1, Emil Lindbäck1, Magne Olav Sydnes1
University of Stavanger1
Bacteria are responsible for millions of deaths per year worldwide and common cures are facing serious challenge due to antibiotic resistance.1 In the past few years, multi-drug resistance bacteria levels have been reported to be increasing significantly, making the development of new solutions very urgent.2
This project, as part of the “Stop Spread Bad Bugs” consortium, aims to provide an answer to the phenomenon of bacteria resistance by using antimicrobial peptides (AMP) and Human milk oligosaccharide (HMOs) activities to overcome the cell membrane barrier and fight bacterial infection more effectively. In this regard, different oligosaccharides were chosen, all of which show antimicrobial and antibiofilm activity,3 with the idea to link them to specific peptides. The perfect candidates for this are non-natural peptide that has a proven antimicrobial activity.
References:
(1) World Health Organization,Ten threats to global health in 2019, 2019.
(2) Baindara, P.; Mandal, S. M. Protein Pept. Lett. 2019, 26, 324-331.
(3) Craft, K. M.; Townsend, S. D. Acc. Chem. Res. 2019, 52, 760−768.
Xuan Thang Nguyen
Emily Martiensen1,2, Xuan Thang Nguyen1,2, Marie Rogne1,2, Synne Lindahl Neeb1,2, Trung Tran4, Sigve Nakken2,3, Bernd Thiede4 and Ragnhild Eskeland1,2
1 Institute of Basic Medical Sciences, Medical Faculty, University of Oslo, Norway,
2 Centre for Cancer Cell Reprogramming (CanCell), Centre of Excellence,
3 Institute of Tumor biology, Oslo University Hospital, Oslo, Norway,
4 Department of Biosciences, University of Oslo, Norway.
Osteosarcoma is a rare and aggressive in terms of bone cancer, and its treatment strategy and high mortality rate have remained unchanged for the past 50 years. Therefore, more knowledge about the disease and its underlying molecular mechanisms is necessary. Transcription factors have been shown to be closely correlated with aggressiveness and response to treatment in osteosarcoma, which highlights the need to investigate the network of transcription factor regulation. In this study, we used a combination of proteomics and transcriptomics analysis to map transcription factor networks in three human osteosarcoma cell lines with different reported aggressiveness. We investigated several master regulators and pioneer transcription factors associated with higher aggressiveness in osteosarcoma, including TP53, SOX, RUNX, and the GATA family’s transcription factors. Furthermore, we established a method to facilitate large-scale chromosome decondensation called lacO-lacI chromatin decompaction assay and CRISPR/Cas9-integrated OsTR1-expressing osteosarcoma cell lines that can be utilized for AID knockdown of transcription factors in functional studies.
Yomki Perez
Yomkippur Perez1,2, Erik Agner1, and Magne Olav Sydnes2
1 Polypure
2 University of Stavanger
Protein and peptide purification has been a major bottleneck in both the pharmaceutical industry and a significant portion of life science research. In all cases, target peptides and proteins must be separated from components of the host cell system or other impurities in chemical synthesis. The well-established gradient chromatography still has issues with column binding and loading capacity. More efficient purification methods are therefore needed.
Displacement chromatography offers an alternative which allows for significantly larger sample loading and more efficient binding. Since the same sample can be processed several times in the column, this method also delivers lower losses. DC has been extensively used in polymer purification and can be translated into peptides and proteins.
Marta Barros
Barros Reguera, Marta1; Mølsæter Maråk, Marte1; Liberg Bergaust, Linda1; Røhr Kjendseth1, Åsmund1 and Sørlie, Morten1
1 NMBU – Norges miljø- og biovitenskapelige universitet
Denitrification is the dissimilatory reduction of nitrate to N2 via nitrite, NO and N2O carried out by a wide range of prokaryotes when oxygen is scarce. Denitrifiers are generally facultative anaerobes, preferentially using oxygen as electron acceptor.
The cd1-type nitrite reductase (NirS) catalyzes the reduction of nitrite to nitric oxide and has also been shown to reduce O2 to H2O in vitro (1), although at significantly lower rate. In vivo experiments with Paracoccus denitrificans indicate that NirS+ cells that switch to aerobic respiration quickly lose the ability to reduce nitrite (2). We hypothesized that NirS is inactivated by oxidative damage, either by presence of reactive oxygen species in the periplasm or by the dual activity of the enzyme.
To explore oxidative damage in vivo, P. denitrificans with a mCherry-nirS gene replacing the native nirS, was exposed to aerobic conditions for variable time-periods. mCherry-NirS was isolated by pull-down using magnetic beads conjugated with mCherry antibodies. Using proteomics, we studied the protein integrity and presence of oxidative damage.
We found that the two histidines (His653 and His465) coordinating the d1 heme iron were prone to suffer oxidation. It is known that His653 is critical in protecting the d1 heme group from oxidative damage (3). The specific and limited localization of the oxidative damage suggests that the loss of NirS activity after aerobic exposures is the result of the dual activity of NirS, which leads to oxidative damage by the formation of reactive oxygen intermediates.
References:
(1) Timkovich R & Robinson MK (1979). Biochem Biophys Res Commun 88, 649–655.
(2) Lycus, P, Soriano-Laguna, M J, Kjos, M, … & Bakken, LR (2018). Proceedings of the National Academy of Sciences, 115(46), 11820-11825.
(3) Centola F, Rinaldo S, Brunori M & Cutruzzolà F. FEBS J. 2006 Oct;273(19):4495-503.
Morten Rese
Morten Rese1, Valerie Schiml1, Magnus Ø. Arntzen1, Anton Stepnov1, Vincent G. H. Eijsink1, Tina R. Tuveng1
1 Faculty of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences (NMBU), P.O. Box 5003, 1432 Ås, Norway
Lignocellulosic biomass is the most abundant renewable carbon source on earth, mainly consisting of cellulose, hemicellulose, and lignin. Enzymatic conversion of cellulose and hemicellulose to fermentable sugars for production of ethanol used in biofuels has gained substantial attention in the past decades. Lignin, however, because of its complex and recalcitrant nature, is usually combusted to generate heat and electricity, despite its potential as a resource for valuable aromatic compounds.
Peroxidases and laccases are redox active enzymes known to be involved in both biosynthesis and degradation of lignin. Lignin-active peroxidases are heme containing enzymes that use hydrogen peroxide to carry out one-electron oxidations in the lignin polymer, which results in modifications such as side‐chain cleavage, demethylation, intramolecular addition, and rearrangements. Laccases are multi copper oxidases that catalyze one-electron oxidations of a broad range of substrates. Catalysis happens at a mononuclear copper site, with subsequent electron transfer to a trinuclear copper site where molecular oxygen binds and is reduced to water.
In this study we elucidate the ability of a recombinantly produced catalase-peroxidase and a multi-copper oxidase to modify lignin. The catalase-peroxidase was identified by meta-omics analysis of anaerobic bacterial enrichments feeding on lignocellulose as the only carbon source, while the multi-copper oxidase is from Cellvibrio japonicus, a well-known lignocellulose degrader. Both enzymes show the ability to oxidize aromatic model compounds.
Diego Sebastian Reyes-Weiss
Diego S. Reyes-Weiss1, Nanna Rhein-Knudsen1, Bjørge Westereng1, Svein J. Horn1
1 Faculty of Chemistry, Biotechnology, and Food Science, Norwegian University of Life Sciences (NMBU), Ås, Norway
Fucoidans are a heterogeneous group of sulfated polysaccharides found in the cell wall of brown seaweeds. Fucoidans have attractive valorisation prospects due to several reported biological activities (Fitton et al. 2019), and recent studies suggest that they play an important role in carbon sequestration (Buck-Wiese et al. 2023). Typically, fucoidans are composed of a linear backbone of α-L-fucose units arranged as α-1,3-, or alternating α-1,3;α-1,4- linkages, with varying degree of sulfation at carbon 2, 3, and 4. Additionally, they can be branched, acetylated, and contain galactose, xylose, mannose, rhamnose, glucose, and uronic acids units. The complex and largely undefined structure of fucoidans, together with their large environmental variability, represent a major challenge in the understanding of their bioactivities and for their processing. Fucoidan-active enzymes, found in many marine bacteria, are classified into different glycosyl hydrolase (GH) families within the carbohydrate-active enzyme database (CAZy). Different types of enzymatic activities have been described, including endo- and exo-fucoidanases, sulfatases, carbohydrate esterases, and lyases, all potential enzymatic tools for processing of fucoidans and for structural elucidation. In this study, we identified a new putative endo-1,3-fucoidanase (FunA50) by blasting the sequence of FunA (Shen et al. 2020), the only characterized member from family GH168, against the genome of Lentimonas sp. CC4, a marine bacterium highly specialized in fucoidan degradation (Sichert et al. 2020). FunA50 was recombinantly produced in E. coli and activity on a fucoidan from Fucus vesiculosus was detected by screening against a collection of fucoidans from 13 brown seaweed species using size-exclusion chromatography. Optimal reaction conditions (35 °C, pH 5.6, and 320 mM NaCl) were determined using the colorimetric PAHBAH method for quantification of reducing ends. Current work is being carried out to elucidate the specificity of FunA50.
References:
(1) Buck-Wiese et al. (2023). Fucoid brown algae inject fucoidan carbon into the ocean. Proc. Nat. Acad. Sci. U.S.A., 120, e2210561119 doi: 10.1073/pnas.2210561119
(2) Fitton et al. (2019). Therapies from fucoidan: New developments. Mar. Drugs, 17, 571. doi: 0.3390/md17100571.
(3) Shen et al. (2020). Discovery and characterization of an endo-1,3-fucanase from marine bacterium Wenyingzhuangia fucanilytica: A novel glycoside hydrolase family. Front. Microbiol., 11:1674. doi: 10.3389/fmicb.2020.01674
(4) Sichert et al. (2020). Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol., 5, 1026-1039. doi: 10.1038/s41564-020-0720-2.
Tobias Rindfleisch
Tobias Rindfleisch1, Jarl Underhaug2 and Markus Miettinen1
1 Department of Chemistry and Computational Biology Unit, University of Bergen, Bergen, Norway
2 Department of Chemistry, University of Bergen, Bergen, Norway
Intrinsically disordered proteins (IDPs) are defined by a lack of a defined 3D structure in aqueous solution. They behave like flexible and strongly dynamic random coils, but demonstrate a characteristic distribution over the conformational space, the so-called structural ensemble. However, IDPs perform important and specific functions at the cellular level and at the super cellular level of the organism.
Molecular dynamics (MD) simulations of IDPs represent a complex problem in bioinformatics, because the most common IDP-specific force fields are not able to describe the dynamics and thus the flexible motions of disordered proteins accurately. In consequence, the development of a force field, which is capable of reproducing the dynamics for IDPs correctly, as confirmed by validation against experimental NMR data, is of special importance.
This project is motivated by the finding of Oh et al. (2012) that dipeptides behave similarly to disordered proteins, suggesting that these elementary building blocks can be used to calibrate an IDP-specific MD model.
A gradient-free evolutionary approach will be used to automatically optimize a previously selected and well performing force field; here the force field parameters are iteratively adjusted to obtain an improved match between the resulting protein dynamics in MD simulations and in NMR experiments (relaxation times, homo and hetero nuclear Nuclear Overhauser Effect). Importantly, the corresponding NMR observables can be computed from the MD simulations directly, without assuming an intervening model. The NMR experiments are performed at the Norwegian NMR Platform.
Keywords: MD simulations, IDPs, NMR relaxation data, Force field development
Greta Daae Sandsdalen
Greta Daae Sandsdalen1, Bjarte Aarmo Lund1, Maryam Imam2, Ole Morten Seternes3, Adele Williamson4 and Hanna-Kirsti Schrøder Leiros1
1 Biomolecular and Structural Chemistry, Department of Chemistry, UiT The Arctic University of Norway
2 The Norwegian College of Fishery Science, UiT The Arctic University of Norway
3 Department of Phamacy, UiT The Arctic University of Norway
4 The University of Waikato, New Zealand
The CRISPR-Cas genome editing system has revolutionized molecular biology, providing an array of biotechnological tools for carrying out precision genome modification and regulation in eukaryotic cells. One limitation of the system at present is that most available tools are developed from and optimized for mesophilic organisms, which limits their utility in extremophilic organisms. Although thermophilic Cas homologs have been developed and verified for use at high temperatures, less attention has been given to the low-temperature end of the spectrum. This paucity of available knowledge on psychrophilic CRISPR-Cas systems and affiliated genome editing tools is particularly problematic for researchers of cold-blooded eukaryotes such as fish biologists, as both the current enzymes and fish cells lose viability at elevated temperatures.
This project is one of three interdisciplinary components of the UiT Strategic-Funded ‘FISH&CRISPR Innovative strategies to improve salmon health’ which aims to establish a platform for the development of a low-temperature CRISPR-Cas genome editing system optimized for salmonids.
In this project, the main goal is to discover and develop one or more CRISPR-associated endonucleases for effective and precise genome editing at low temperatures. A second aim is to investigate the abundance of CRISPR-systems across cold-adapted bacteria through bioinformatics analysis.
Through the analysis of 952 genomes of cold-adapted bacteria, it has been found that CRISPR-Cas systems are present in low abundance, with only 20% of the genomes containing them. Among the identified CRISPR-Cas systems, Class 1 is the most frequently observed. However, Type II-C with the Cas9 endonuclease is the dominant Class 2 CRISPR-Cas system. From five unique psychrophilic bacteria, five Cas endonucleases has been expressed recombinantly in soluble form. Out of the five, three has been purified at low concentrations and is currently being characterized.

Bruna Schuck
Bruna Schuck1,2, Raphael Klein3, Vipul Panchal1, Ruth Brenk1,2
1Department of Biomedicine, University of Bergen, Bergen, Norway,
2MepAnti Consortium, 3BioSolveIT, Sankt Augustin, Germany
Antibiotic resistance looms as a serious threat to our well-being, health care systems and, consequently, the global economy as we continue to overuse and misuse antibiotics (1). Therefore, the discovery and development of new antibiotics are urgently needed. The 2C-methyl-D-erythritol 4-phosphate (MEP) pathway generates precursors of terpenoids, natural products essential for many human pathogens (2). As this pathway is absent in humans, its enzymes are attractive targets for new antibiotics.
In this project, we target 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR), the second enzyme of the MEP pathway, from Pseudomonas aeruginosa (PaDXR). Two main routes were chosen to find new inhibitors: an experimental route using Bio-Layer Interferometry (BLI) and a computational route using Chemical Space Docking. Using BLI, a fragment screening of our in-house carboxylic acids’ library (302 fragments) was performed to find potential binders to PaDXR. Following a single point binding assay, a serial dilution of selected fragments led to a few measured KDs. Currently, the same library is being screened with DXR’s coenzyme, NADPH, in the buffer.
A novel approach for virtual screening, Chemical Space Docking, was carried out for Escherichia coli DXR (EcDXR) in collaboration with BioSolveIT, Germany. Four chemical spaces (virtual combinatorial libraries), representing over 7 trillion virtual product molecules, were utilized: a custom space based on Specs’ building blocks and robust organic reactions, Enamine’s REAL space, eMolecules’ eXplore space and WuXi’s galaXi space. First, the building blocks of each space were transformed into fragments based on the given reactions. Only those with one linker, which would allow the enumeration of said fragment afterward, were taken into account.
The initial fragments were docked with FlexX 5.2.0 and scored with Hyde Scorer 1.5.0 and, upon their affinity scores and visual inspection, the most promising fragments’ poses were chosen to undergo enumeration. A total of 336 fragments were chosen and a sublibrary of their possible virtual products was generated for each initial fragment. Next, template-based docking and scoring were done for those virtual products, again with FlexX and Hyde, using the fragments’ poses chosen as templates. The filtering and selection of a set of compounds is ongoing. Shortlisted protein-ligand complexes will be simulated with YASARA, before purchasing and/or synthesizing some of those for in vitro testing.
References:
(1) Murray, C. J. et al (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet, 399(10325), 629–655.
(2) Gräwert, T. et al (2011). Biochemistry of the non-mevalonate isoprenoid pathway. Cellular and Molecular Life Sciences, 68(23), 3797–3814.
Rannei Skaali
Rannei Skaali, Dag Ekeberg, Hanne M. Devle, and Morten Sørlie
Faculty of Chemistry, Biotechnology, and Food Science, Norwegian University of Life Sciences, Ås, Norway
Lignin is abundant in nature and can be biorefined by reductive catalytic fractionation (RCF) for industrial use. Monolignols such as 4-propylphenol, 4-propylguaiacol, and 4-propylsyringol are obtained from lignin in this process. The commercial value of monolignols is increased by monolignol oxyfunctionalization that can be used in a sustainable green industry. Using synthetic chemicals to oxidize monolignols is not sustainable due to their environmental impact, costs, and low selectivity. However, enzymes are a green alternative and unlike synthetic chemicals, can catalyze selective oxyfunctionalization. Preferable positions for monolignol oxidation are the Ca and Cg producing the corresponding benzoic acid and alcohol, aldehyde, or carboxylic acid, respectively (Fig. 1). Here, we present a validated method for the identification and quantification of native and oxidized monolignols by direct infusion electrospray ionization mass spectrometry (DI-ESI-MS).

Figure 1: Native and oxidized monolignols. Monolignols such as 4-propylphenol (R = R’ = H), 4-propylguaiacol (R = H, R’ = OCH3), and 4-propylsyringol (R = R’ = OCH3) are obtained from lignin. Pseudomolecular ions are observed in a total ion chromatogram (MS) which can be differentiated from one another by fragmentation using collision-induced dissociation (MS2).
Amalie Skogvold
Amalie Skogvold1, Heidi Therese Hillier1, Heidi Erlandsen1, Ingar Leiros1
1 Department of Chemistry, UiT – The Arctic University of Norway
ω-transaminases are highly sought after biocatalysts in the pharmaceutical industry, as a greener alternative for production of valuable chiral amines in enantiopure form. The DABA ω-transaminases EctB from the bacterial Ectoine biosynthesis pathway catalyzes the transamination of L-aspartic-4-semialdehyde (ASA) using glutamic acid as an amino donor, converting them to 1,4-diaminobutyric acid (DABA) and 2-oxoglutaric acid, respectively. Ectoine is a highly valuable compound used both in the cosmetics and pharmaceutical industry, due to its many novel properties such as: cell protectant, protein stabilizer, skin and UV protectant. It is produced mainly by a method called bacterial milking, but more recent research is done on heterologous protein production using non-halophilic bacteria such as Escherichia coli. In the Ectoine Research group, we aim to characterize the core enzymes in the Ectoine pathway, including EctA, EctB, EctC and additionally the transaminase DoeD from the degradation pathway. Previous work includes solving the crystal structure of EctB from the model organism for Ectoine production Chromohalobacter salexigens (Hillier et al., 2020). The first crystal structure and biochemical characterization of DoeD from the same bacteria has also been finalized.
Recently, we solved the first crystal structure of EctB from a psychrophilic and halophilic bacterium and completed a biochemical and biophysical characterization of the transaminase, including exploring the substrate scope for other potential uses of the enzyme as a pharmaceutical biocatalyst. Based on the structural analysis, we have designed mutants to validate residues important for activity and to modulate substrate specificity, flexibility, and PLP binding for increased enzyme life-time. There is ongoing work on the productionand testing of the mutants in comparison to the wildtype, including activity assays, substrate scope, SEC-MALS (oligomeric state) and DSC (protein stability).
References:
(1) Hillier, H. T., Altermark, B., & Leiros, I. (2020). The crystal structure of the tetrameric DABA‐aminotransferase EctB, a rate‐limiting enzyme in the ectoine biosynthesis pathway. The FEBS journal, 287(21), 4641-4658.
Anita Solem
Anita Solem1, Agnes B. Petersen1, Anne Tøndervik2, Finn Lillelund Aachmann1
1 Department of Biotechnology and Food Science, NTNU Norwegian University of Science and Technology, Trondheim, Norway
2 Department of Biotechnology and Nanomedicine, SINTEF Industry, Trondheim, Norway.
The biopolymer alginate, produced by brown algae and a few bacterial species, consists of the monomers β-d-mannuronic acid (M) and α-l-guluronic (G) acid arranged in blocks of only M, only G or alternating M and G units. Alginate has a range of industrial and medical applications, depending on the polymer’s composition and length. G-oligomers has shown to be effective in disrupting biofilms and aiding the diffusion of antimicrobial agents through mucus layers, and thus have potential in the treatment of diseases such as cystic fibrosis.
Alginate epimerases and alginate lyases are enzymes that can modify alginates by introducing G-residues and cleaving the alginate chain, respectively. Such modifications may make the alginate more suitable for industrial and medical applications. Genetic modification of native alginate epimerases and lyases may enhance their ability to create valuable alginate products and enable enzymatic G-oligomer production. The bifunctional alginate epimerase/lyase AlgE7 produces short oligomers containing both M- and G-residues. We created chimeric enzymes by inserting the active module of AlgE7 into the alginate epimerase AlgE1. These chimeric enzymes have the potential to generate G-oligomers of a specific length, which is both medically and industrially relevant. The chimeric enzymes were characterized by spectrophotometric assays, nuclear magnetic resonance (NMR) and high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD).
The chimeric enzymes presented as different from the native AlgE1 and AlgE7, both in product profile and catalytic activity. One of the enzymes showed increased G-block content when acting on poly-M, as compared to AlgE7. As a result, we believe that combining alginate epimerases with the active module of an alginate lyase is a good starting point for developing novel enzymes capable of synthesizing valuable alginate products such as G-oligomers.
Thomas Stevenson
Thomas Stevenson1, Jack Law1, Evan Spruijt3, Sushma-Nagaraja Grellscheid1,2
1 Computational Biology Unit, Department of Biological Sciences, University of Bergen, Norway
2 Department of Biosciences, University of Durham, United Kingdom
3 Institute for Molecules and Materials, Radboud University, Nijmegen, The Netherlands
Biomolecular condensates are highly organized and dynamic assemblies distinct from classical organelles or protein complexes. They are liquid-like and possess a biochemically discrete composition maintained without a traditional membrane. Stress granules (SGs) are an example of a biomolecular condensate that forms in response to cellular stressors such as oxidative or heat stress or the accumulation of misfolded proteins. SGs serve to rapidly reprogramme translation by sequestering protein, mRNA, and ribosomal subunits to promote cellular survival, releasing their content once the stress has resolved. This profound role in cellular physiology is implicated in various cellular operations, including disease processes such as cancer, viral infection, and neurodegenerative disorders. For example, solid-like protein inclusions in age-related neurodegenerative disorders such as Alzheimer’s and Huntington’s disease share many of the same intrinsically disordered proteins that comprise
SGs and other condensates. Thus, SGs may exist as metastable liquid-like phases with the potential to transition into pathological solid-like assemblies if the antecedent conditions exist. Understanding the principles behind the formation of biomolecular condensates is essential for developing strategies and therapeutics that modify the properties of stress granules. Here we have investigated how ammonium ions disrupt RNA-mediated condensation and prevent the assembly of stress granules and other condensates in vivo. Our data highlight how the cellular environment can modulate condensates’ physical and biological properties/functions and how this may be implicated in disease processes.
Szymon Mikolaj Szostak
Nico König1,2, Szymon Mikołaj Szostak1, Josefine Elisø Nielsen1, Martha Dunbar3, Su Yang4, Weike Chen4, Ari Benjamin3, Aurel Radulescu2, Najet Mahmoudi5, Lutz Willner2, Sinan Keten3, He Dong4, Reidar Lund1
1 Department of Chemistry, University of Oslo, Oslo, Norway,
2 Jülich Centre for Neutron Science, Jülich, Germany
3 Department of Mechanical Engineering, Northwestern University, Evanston, United States
4 Department of Chemistry & Biochemistry, The University of Texas at Arlington, Arlington, United States
5 ISIS-STFC, Rutherford Appleton Laboratory, Chilton, United Kingdom
Often nanostructures formed by small molecules and based on hydrophobic interactions are rather unstable, with the tendency to dissolution and molecular exchange in aqueous media.1 Contrary to them, peptides offer precise control over nanostructures’ morphology through a variety of interactions and the possibility to increase stability via rational design.
In the research, we investigated a family of peptides forming beta-sheet nanofibers.2 We employed small angle X-ray/neutron scattering, circular dichroism and molecular dynamics techniques to investigate detailed structure and stability. The results give important insight into the relationship between molecular structure and nanofibers stability, which is important for designing peptides for biomedical applications.3

References:
(1) Choi, S. H., Bates, F. S. & Lodge, T. P. Molecular exchange in ordered diblock copolymer micelles. Macromolecules 44, 3594–3604 (2011).
(2) Dong, H., Paramonov, S. E., Aulisa, L., Bakota, E. L. & Hartgerink, J. D. Self-assembly of multidomain peptides: Balancing molecular frustration controls conformation and nanostructure. J. Am. Chem. Soc. 129, 12468–12472 (2007).
(3) Sunna, A., Care, A. & Bergquist, P. L. Peptides and Peptide-based Biomaterials and their Biomedical Applications. Advances in Experimental Medicine and Biology (Springer International Publishing AG, 2017).
Mary Dayne Sia Tai
Mary Dayne S. Tai1, Marte I. Flydal1, Jorge Cuéllar2, Lorea Velasco3, Lissette Ochoa2, Kunwar Jung-KC1, Juha P. Kallio1, Svein I. Støve1, Arturo Muga3, José María Valpuesta2 and Aurora Martinez1
1 Department of Biomedicine, University of Bergen, 5009 Bergen, Norway
2 Centro Nacional de Biotecnología (CNB-CSIC), Madrid, Spain
3 Instituto Biofisika (UPV/EHU, CSIC), Universidad del País Vasco, (UPV/EHU), and Departamento de Bioquímica y Biología Molecular, Facultad de Ciencia y Tecnología, Universidad del País Vasco, (UPV/EHU), Barrio Sarriena, 48940 Leioa, Spain
The Hsp40 chaperone protein DNAJC12 binds to tyrosine hydroxylase (TH), the rate-limiting enzyme in the synthesis of catecholamine neurotransmitters dopamine, adrenaline and noradrenaline. Pathogenic variants in DNAJC12 are associated with early-onset parkinsonism, and other symptoms associated with TH deficiency (THD), but the lack of biophysical, biochemical and structural studies on full-length DNAJC12 and the TH:DNAJC12 complex have so far hindered the understanding of the function of the TH:DNAJC12 complex, as well as the pathogenic mechanisms behind DNAJC12 disease variants. In this work we show that DNAJC12 binds to TH and forms a stable complex amenable for further characterization. By using cryo-electron microscopy and cross-linking mass spectrometry, we have obtained structural data on the complex, which together with the biochemical characterization confirm the molecular determinants for DNAJC12 binding to TH and demonstrate the molecular disease mechanisms of DNAJC12 variants found in patients.
Reza Talandashti
Reza Talandashti1, Mahmoud Moqadam1, Charlotte Gehin2, Anne-Claude Gavin3, Nathalie Reuter1
1 Computational Biology Unit, Department of Chemistry, University of Berge, Bergen, Norway
2 École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland.
3 Department of Cell Physiology and Metabolism, Faculty of Medicine, University of Geneva, Geneva, Switzerland.
STARD4 (steroidogenic acute regulatory protein-related lipid-transfer domain 4) is a START domain with non-vesicular cholesterol transfer activity from the plasma membrane (PM) to the endoplasmic reticulum (ER) membrane. The lipid compositions of the plasma and ER membranes are considerably different, yet STARD4 is able to selectively bind to the PM, extract a cholesterol molecule into its hydrophobic pocket, which it subsequently delivers to the ER membrane following adsorption on the ER. While structural elements involved in STARD4 membrane binding site have been suggested, the mechanism for membrane selectivity and associated conformational dynamics have remained unclear. Herein, we report the results of μs-long all-atom molecular dynamics (MD) simulations and experiments to propose a mechanism for the membrane selectivity of STARD4. We found that the orientation of STARD4 at the PM is dependent on the presence of phosphoinositide (PIP2) lipids. We identify a specific PIP2 binding site on β1/β2 strands. The absence of PIP2 lipids in PM leads to a secondary binding mode with a different binding orientation where the cavity is not oriented properly to allow lipid extraction. In agreement with our findings, the experimental data shows increased binding of STARD4 to PIP2-containing vesicles. Additionally, our simulations captured the conformational change of the Ω1 loop in the holo STARD4, and this determines the binding mode of STARD4 to the ER membrane. We observed that in addition to the movement of the Ω1 loop, the flexibility of the Ω4 loop is essential to close and open the gate of the binding pocket. Overall, these findings suggest that the ability of STARD4 to selectively bind to the PM and the ER membrane is controlled by the lipid composition and the state of the protein.
Juliana Miranda Tatara
Juliana M Tatara1, Amanda Moraes1, Walter O. Beys-da-Silva1,2, Lucélia Santi1,2
1 Post-Graduation Program in Cellular and Molecular Biology, UFRGS. Porto Alegre, RS. Brazil.
2 Center of Experimental Research, Clinical Hospital of Porto Alegre. Porto Alegre, RS. Brazil
Klebsiella pneumoniae is a gram-negative encapsulated bacterium part of the ESKAPE group, which comprises six highly virulent and antibiotic resistant bacterial pathogens [1]. While most microorganisms are unable to resist the microbicidal effect of human serum, some serum-resistant bacteria have evolved and developed mechanisms to subvert this barrier [2-3]. Although efforts have been made to understand the mechanisms involved in serum resistance, the molecular mechanisms are still poorly understood. Herein, we use proteomics (MALDI-TOF mass spectrometry) to evaluate the molecular alterations in a clinical isolated strain of K. pneumoniae (resistant to 16 antibiotics) triggered after active and heat-inactivated serum (control) exposure for 1 and 4 hours, and its potential antibacterial targets. Cell viability assay indicates K. pneumoniae to be resistant to active serum. After one hour of active serum exposure, “thiamine biosynthetic process” was the third most significant impacted biological process and KEGG analysis pointed to a modulation of “thiamine metabolism”, highlighting 4 upregulated proteins in this metabolic pathway. This suggests a higher production of thiamine diphosphate/pyrophosphate (TPP), an essential cofactor for several cellular processes [4] which is also involved in the pathogenesis of Pseudomonas aeruginosa [5]. In situations of nutritional deprivation or a stressful environment, Escherichia coli can accumulate thiamine as a defence mechanism [6]. When we added exogenous thiamine to active serum, bacteria presented increased resistance. Overall, our data suggest that K. pneumoniae changes its metabolism to evade the host immune system, as can be observed in the predicted activation of the “thiamine metabolism”, specifically the production of TPP metabolite. Further experiments must be conducted to explore this relationship with serum resistance, as well as to develop novel therapeutic targets, such as inactivation of some enzymes within the Thiamine Metabolism pathway.
(1) De Oliveira, David M P et al. “Antimicrobial Resistance in ESKAPE Pathogens.” Clinical microbiology reviews vol. 33,3 e00181-19. 13 May. 2020, doi:10.1128/CMR.00181-19
(2) Samant, Shalaka et al. “Nucleotide biosynthesis is critical for growth of bacteria in human blood.” PLoS pathogens vol. 4,2 (2008): e37. doi:10.1371/journal.ppat.0040037
(3) Sheldon, Jessica R et al. “Iron Acquisition Strategies of Bacterial Pathogens.” Microbiology spectrum vol. 4,2 (2016): 10.1128/microbiolspec.VMBF-0010-2015. doi:10.1128/microbiolspec.VMBF-0010-2015
(4) Bettendorff, Lucien et al. “Thiamine triphosphate: a ubiquitous molecule in search of a physiological role.” Metabolic brain disease vol. 29,4 (2014): 1069-82. doi:10.1007/s11011-014-9509-4
(5) Kim, Hyung Jun et al. “The ThiL enzyme is a valid antibacterial target essential for both thiamine biosynthesis and salvage pathways in Pseudomonas aeruginosa.” The Journal of biological chemistry vol. 295,29 (2020): 10081-10091. doi:10.1074/jbc.RA120.013295
(6) Lakaye, Bernard et al. “Thiamine triphosphate, a new signal required for optimal growth of Escherichia coli during amino acid starvation.” The Journal of biological chemistry vol. 279,17 (2004): 17142-7. doi:10.1074/jbc.M313569200
Susanne Hansen Troøyen
Susanne Hansen Troøyen1, Gaston Courtade1
Department of Biotechnology and Food Science, NTNU Norwegian University of Science and Technology, Trondheim, Norway
Carbohydrate-binding modules (CBMs) are essential for binding of carbohydrate-active enzymes to insoluble substrates. However, many details about the binding mechanism of CBMs to their substrates remains unclear. Here, we investigate how a CBM (ScCBM2 from Streptomyces coelicolor1,2) binds to cellulose and polyethylene terephthalate (PET) substrates using NMR spectroscopy and spectrophotometry. We explore how the structure of the binding site affects binding affinity by site-directed mutagenesis of specific amino acids, and how differences in substrate structures affect both affinity and binding capacity. This gives more information about the binding mechanism of CBMs, which could help to develop new methods for enzymatic depolymerization of cellulose and PET.
References:
(1) Courtade, G.; Forsberg, Z.; Vaaje-Kolstad, G.; Eijsink, V. G. H.; Aachmann, F. L. Chemical Shift Assignments for the Apo-Form of the Catalytic Domain, the Linker Region, and the Carbohydrate-Binding Domain of the Cellulose-Active Lytic Polysaccharide Monooxygenase ScLPMO10C. Biomolecular NMR Assignments 2017, 11 (2), 257–264. https://doi.org/10.1007/s12104-017-9759-2.
(2) Courtade, G.; Forsberg, Z.; Heggset, E. B.; Eijsink, V. G. H.; Aachmann, F. L. The Carbohydrate-Binding Module and Linker of a Modular Lytic Polysaccharide Monooxygenase Promote Localized Cellulose Oxidation. Journal of Biological Chemistry 2018, 293 (34), 13006–13015. https://doi.org/10.1074/jbc.RA118.004269.
Rosaliina Turunen
Rosaliina Turunen1, Tina R. Tuveng1, Valerie Schiml1, Vincent GH. Eijsink1, Magnus Ø. Arntzen1
1 Faculty of Chemistry, Biotechnology and Food Science, Norwegian University of Life Sciences, Ås, Norway
Background: Freshwater ecosystems are largely influenced by neighboring agriculture fields where fertilizer run-off may leak into surrounding water bodies. To counter-act this eutrophic driver, farmers utilize water treatment basins filled with woodchips in which microorganisms convert nitrogen into nitrogen-gases, while degrading lignocellulose. While polysaccharide-degrading strategies have been well-described for aerobic and anaerobic microorganisms, this is largely unknown for denitrifying microorganisms.
Within these habitats, we have discovered Cellulomonas gelida, a denitrifying, cellulose degrader encoding two novel enzymes belonging to the LPMO protein family. One of these is expressed in anoxia, rising key questions about this enzyme’s function and source of O2. LPMOs catalyze the oxidative cleavage of polysaccharides under aerobic conditions either with oxygen (O2) or in anoxia if provided with hydrogen peroxide (H2O2). To the best of our knowledge, this is the first time an LPMO has been detected under anaerobic conditions.
Experimental: The two LPMOs were expressed in E. coli BL21, purified by anion exchange and size exclusion chromatography, and characterized for substrate and regiospecificity using MALDI and high-performance anion exchange chromatography (HPAEC).
Results: CgLPMO10B was able to oxidize cellulose (Avicel and PASC), and appeared to be strictly C1-oxidizing, while no activity could be observed towards xylan or chitin. CgLPMO10A showed no activity towards any of the tested substrates.
Using multiple isolation steps, C. gelida was isolated from the enrichment cultures and identified using 16S rRNA sequencing. To explore the relationship of the isolated strain to known strains of the Cellulomonas genus, whole genome sequencing will be conducted, as well as growth experiments to elucidate LPMO expression under various conditions (denitrifying, micro-oxic, oxic).
These results provide a great start for further characterization of CgLPMO10B.
Gabriela Ruiz Velarde
Ruiz-Velarde Gabriela1, Lopez-Moreno Andrea Johana1, Kalienkova Valeriia1, Kursula Petri1
1 Department of Biomedicine, University of Bergen, Jonas Lies Vei 91, 5009, Bergen, Norway
The myelin sheath is a specialized layer wrapped around the nerve axons, which comprises lipid bilayers and specific proteins that participate in the stacking of the membrane. Both lipids and proteins form nanoclusters to form a biologically functional proteolipid membrane multilayer. While myelin membranes are rich in protein content, only a few specific proteins are found in these membranes. P0 is the most abundant protein in the Peripheral Nervous System and the proteolipid protein (PLP) and its spliced form, DM20, comprise 17 to 45% of the total compact myelin protein in the Central Nervous System. PLP in particular has been linked with some neuropathies, such as Pelizaeus-Merzbacher disease (PMD) and spastic paraplegia type 2.
PLP belongs to the Tetraspan protein family, it is embedded in the membrane and has four hydrophobic transmembrane helices. Even though PLP has been extensively studied, no high-resolution structures of this protein are available, its small size makes it difficult to apply Cryo-EM to study the structure and the fact that it’s embedded in the membrane could result in a low yield of the expression and purification of the protein to obtain crystals. In addition, little is known about the lipid interaction properties and complex formation with other myelin proteins, and how they organize to form the myelin membrane.
The project aims to address these questions using a combination of biophysical techniques. Protein scaffolds will be implemented in order to increase the size and stability of the proteins and thereby enable structure determination using single-particle cryo-EM. Additionally, once high-resolution data is obtained, the dynamics of PLP in multi-layered biomimetic membranes are going to be studied by Synchrotron CD spectroscopy (SRCD) and Small Angle X-ray Scattering (SAXS), as well as computer simulations. Novel insights will be elucidated, such as the structure of PLP and protein-membrane interactions. Understanding the dynamics and the formation of large-scale structures will be helpful in better-comprehending myelin formation.
Amanda Kristine Votvik
Amanda K. Votvik1, Åsmund K. Røhr1, Bastien Bissaro2, Anton A. Stepnov1, Morten Sørlie1, Vincent G. H. Eijsink1 and Zarah Forsberg1
1 Faculty of Chemistry, Biotechnology, and Food Science, The Norwegian University of Life Sciences (NMBU), 1432 Ås, Norway.
2 INRAE, Aix Marseille University, UMR1163 Biodiversité et Biotechnologie Fongiques, 13009, Marseille, France
Bacterial lytic polysaccharide monooxygenases (LPMOs) oxidize abundant and recalcitrant polymers such as chitin and cellulose. Interestingly, the genome of the model actinomycete Streptomyces coelicolor A3(2) encodes seven putative LPMOs. Phylogenetic analysis shows that four of these putative LPMOs group with typical chitin-active LPMOs, two with typical cellulose-active LPMOs, whereas one stands out by being part of a novel subclade of non-characterized enzymes. The latter enzyme, called ScLPMO10D, stands out among S. coelicolor LPMOs as the only featuring a C-terminal extension with a cell wall sorting signal (CWSS), which is expected to covalently anchor the enzyme to the cell wall peptidoglycan layer. Here, we have produced a truncated version of ScLPMO10D without the CWSS and determined its crystal structure, EPR spectrum, and various functional properties. Although showing several structural and functional features typical for cellulose-active LPMO10s, ScLPMO10D showed oxidative activity towards chitin only. Comparison with two other chitin-active LPMO10s of bacterial origin revealed interesting functional differences related to the reactivity of the catalytic copper ion. The characterization of this unique LPMO sheds light on structure-function relationships in phylogenetically distant LPMOs with similar substrate specificities and adds to our understanding of how structural variation contributes to fine-tuning of LPMO function.
Ayla Coder
Ayla Coder
Faculty of Medicine, University of Oslo, Oslo, Norway,
The pathogenic bacteria Vibrio cholerae cause millions of cholera infections and hundreds of thousands of recorded deaths globally each year. There is an adhesin used by V. cholerae to colonize the small intestines of mammals, and to adhere to chitin in plankton and shellfish. This adhesin has lytic polysaccharide monooxygenase (LPMO) activity, which can break down complex carbohydrates. The molecular interaction of this LMPO with the bacteria is however poorly understood. The aim of this project is to determine the mechanism by which this adhesin binds to chitin. A combination of different methods will be used, including binding assays, EM, cryo-EM, and NMR.
A deeper understanding of the molecular mechanisms is of interest in more than one way. They may inform new treatments for cholera or other infections, and may even lead to improved ways for biofuel production.









































